Project
DANIO-CODE
Navigation
Downloads
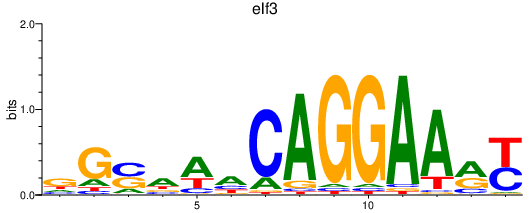
Results for elf3
Z-value: 1.15
Transcription factors associated with elf3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
elf3
|
ENSDARG00000077982 | E74-like factor 3 (ets domain transcription factor, epithelial-specific ) |
Activity profile of elf3 motif
Sorted Z-values of elf3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of elf3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| KN150589v1_-_5209 | 3.04 |
ENSDART00000157761
ENSDART00000157531 |
elovl7b
|
ELOVL fatty acid elongase 7b |
| chr17_+_19606608 | 2.96 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr6_+_3556296 | 2.75 |
ENSDART00000041627
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr25_+_4888647 | 2.69 |
ENSDART00000165933
ENSDART00000159537 |
hdac10
|
histone deacetylase 10 |
| chr11_-_41751151 | 2.67 |
ENSDART00000173086
|
zgc:175088
|
zgc:175088 |
| chr23_+_32174669 | 2.36 |
ENSDART00000000992
|
ENSDARG00000000887
|
ENSDARG00000000887 |
| chr22_-_17627900 | 2.34 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr19_-_20819477 | 2.15 |
ENSDART00000151356
|
dazl
|
deleted in azoospermia-like |
| chr13_-_37340209 | 2.11 |
|
|
|
| chr18_+_27769446 | 1.99 |
|
|
|
| chr6_-_10492276 | 1.95 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr22_-_17627831 | 1.94 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr19_-_35400339 | 1.85 |
|
|
|
| chr10_-_180603 | 1.83 |
|
|
|
| chr12_+_20505619 | 1.80 |
ENSDART00000074384
|
stx4
|
syntaxin 4 |
| chr5_-_28931727 | 1.74 |
ENSDART00000174697
|
arrdc1a
|
arrestin domain containing 1a |
| chr17_-_44326992 | 1.70 |
ENSDART00000148786
|
exoc5
|
exocyst complex component 5 |
| chr5_-_56641212 | 1.68 |
ENSDART00000050957
|
fer
|
fer (fps/fes related) tyrosine kinase |
| chr23_-_32267833 | 1.64 |
|
|
|
| chr20_-_13729816 | 1.63 |
ENSDART00000078893
|
ezrb
|
ezrin b |
| chr21_+_6008781 | 1.59 |
ENSDART00000141607
|
fpgs
|
folylpolyglutamate synthase |
| chr14_-_21779536 | 1.57 |
|
|
|
| chr19_+_43745546 | 1.55 |
ENSDART00000145846
|
sesn2
|
sestrin 2 |
| chr23_+_32175042 | 1.54 |
ENSDART00000138849
|
ENSDARG00000000887
|
ENSDARG00000000887 |
| chr16_-_15497269 | 1.51 |
ENSDART00000053754
|
has2
|
hyaluronan synthase 2 |
| chr19_+_43745687 | 1.50 |
ENSDART00000102384
|
sesn2
|
sestrin 2 |
| chr14_-_6901209 | 1.50 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr5_-_56641061 | 1.50 |
ENSDART00000050957
|
fer
|
fer (fps/fes related) tyrosine kinase |
| chr3_+_479366 | 1.48 |
|
|
|
| chr25_-_19889225 | 1.44 |
|
|
|
| chr13_-_35782121 | 1.43 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr14_-_27040866 | 1.41 |
ENSDART00000173053
|
CU062454.1
|
ENSDARG00000105327 |
| chr11_-_41751050 | 1.40 |
ENSDART00000173086
|
zgc:175088
|
zgc:175088 |
| chr2_-_24413154 | 1.38 |
ENSDART00000145526
|
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr5_-_23741221 | 1.37 |
ENSDART00000042481
|
phf23a
|
PHD finger protein 23a |
| chr20_-_34126039 | 1.36 |
ENSDART00000033817
|
scyl3
|
SCY1-like, kinase-like 3 |
| chr8_+_3373066 | 1.34 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr17_+_44327269 | 1.32 |
ENSDART00000045882
|
ap5m1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr6_-_44164161 | 1.30 |
ENSDART00000035513
|
shq1
|
SHQ1, H/ACA ribonucleoprotein assembly factor |
| chr15_-_38027363 | 1.29 |
ENSDART00000157550
|
si:dkey-238d18.15
|
si:dkey-238d18.15 |
| chr10_-_25383262 | 1.28 |
ENSDART00000166348
|
cct8
|
chaperonin containing TCP1, subunit 8 (theta) |
| chr5_-_40524177 | 1.28 |
ENSDART00000083561
|
mtmr12
|
myotubularin related protein 12 |
| chr5_-_56641001 | 1.26 |
ENSDART00000149855
|
fer
|
fer (fps/fes related) tyrosine kinase |
| chr20_-_18482817 | 1.26 |
ENSDART00000159636
|
vipas39
|
VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| chr2_-_30216377 | 1.26 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr5_-_38570658 | 1.25 |
ENSDART00000020808
|
paqr3a
|
progestin and adipoQ receptor family member IIIa |
| chr3_+_48810347 | 1.25 |
ENSDART00000133283
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr13_-_21541531 | 1.24 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr7_+_56076854 | 1.22 |
|
|
|
| chr23_+_26099718 | 1.22 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr14_+_31596468 | 1.21 |
ENSDART00000173259
|
BX005454.1
|
ENSDARG00000100646 |
| chr6_-_10492123 | 1.21 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr16_-_29755471 | 1.20 |
ENSDART00000133965
|
scnm1
|
sodium channel modifier 1 |
| chr4_-_17364090 | 1.20 |
ENSDART00000134467
|
parpbp
|
PARP1 binding protein |
| chr1_+_32825950 | 1.18 |
ENSDART00000018472
|
chmp2bb
|
charged multivesicular body protein 2Bb |
| chr10_-_180681 | 1.18 |
|
|
|
| chr5_+_35186037 | 1.17 |
ENSDART00000127383
ENSDART00000022043 |
stard8
|
StAR-related lipid transfer (START) domain containing 8 |
| chr19_+_4135556 | 1.16 |
ENSDART00000162461
|
btr26
|
bloodthirsty-related gene family, member 26 |
| chr24_+_25995817 | 1.16 |
ENSDART00000139017
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr5_-_56640932 | 1.15 |
ENSDART00000149855
|
fer
|
fer (fps/fes related) tyrosine kinase |
| chr20_-_33802082 | 1.13 |
ENSDART00000166573
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr5_-_38571198 | 1.12 |
ENSDART00000020808
|
paqr3a
|
progestin and adipoQ receptor family member IIIa |
| chr6_+_13766964 | 1.12 |
ENSDART00000136006
ENSDART00000009382 |
dnpep
|
aspartyl aminopeptidase |
| chr18_+_45669615 | 1.11 |
ENSDART00000150973
|
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr5_+_40235387 | 1.10 |
ENSDART00000147767
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr19_+_43745515 | 1.09 |
ENSDART00000145846
|
sesn2
|
sestrin 2 |
| chr8_+_150970 | 1.08 |
|
|
|
| chr10_-_44173041 | 1.06 |
|
|
|
| chr4_-_7861030 | 1.06 |
ENSDART00000067339
|
mcm10
|
minichromosome maintenance 10 replication initiation factor |
| chr1_+_157840 | 1.05 |
ENSDART00000152205
ENSDART00000160843 |
cul4a
|
cullin 4A |
| chr4_+_17653548 | 1.02 |
ENSDART00000026509
|
cwf19l1
|
CWF19-like 1, cell cycle control |
| chr16_+_38251653 | 1.01 |
ENSDART00000044971
|
myo1eb
|
myosin IE, b |
| chr16_+_38188364 | 0.98 |
ENSDART00000131694
|
selenbp1
|
selenium binding protein 1 |
| chr4_-_12979905 | 0.97 |
ENSDART00000150674
|
tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr15_-_5810906 | 0.97 |
ENSDART00000142334
|
hnrnpl2
|
heterogeneous nuclear ribonucleoprotein L2 |
| chr22_-_16351654 | 0.96 |
ENSDART00000168170
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr20_-_22899048 | 0.96 |
ENSDART00000063609
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr2_-_30216333 | 0.96 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| KN149781v1_+_5174 | 0.96 |
ENSDART00000165339
|
CDC37
|
cell division cycle 37 |
| chr8_+_3372903 | 0.95 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr16_-_41718971 | 0.94 |
|
|
|
| chr12_-_26445609 | 0.94 |
ENSDART00000132737
ENSDART00000163931 |
acsf2
|
acyl-CoA synthetase family member 2 |
| chr8_+_50754290 | 0.93 |
ENSDART00000155664
|
si:ch73-6l19.2
|
si:ch73-6l19.2 |
| chr17_+_24045582 | 0.93 |
ENSDART00000132755
|
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr16_+_29574449 | 0.92 |
ENSDART00000148450
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr24_+_16575167 | 0.91 |
|
|
|
| chr19_-_24541229 | 0.90 |
ENSDART00000080632
|
zgc:64022
|
zgc:64022 |
| chr5_-_45986774 | 0.90 |
|
|
|
| chr4_-_12979951 | 0.90 |
ENSDART00000013839
|
tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr10_-_20524937 | 0.90 |
|
|
|
| chr24_-_13205000 | 0.90 |
ENSDART00000134482
|
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr2_-_27996140 | 0.89 |
ENSDART00000097868
|
tgs1
|
trimethylguanosine synthase 1 |
| chr20_+_2100836 | 0.89 |
|
|
|
| chr13_-_33348231 | 0.89 |
ENSDART00000160520
|
mad2l1bp
|
MAD2L1 binding protein |
| chr8_-_47339136 | 0.87 |
ENSDART00000060853
|
pex10
|
peroxisomal biogenesis factor 10 |
| chr12_+_30590876 | 0.86 |
ENSDART00000066259
|
kcnk1a
|
potassium channel, subfamily K, member 1a |
| chr14_+_21779325 | 0.85 |
ENSDART00000106147
|
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr15_-_35242832 | 0.84 |
ENSDART00000099677
|
bloc1s3
|
biogenesis of lysosomal organelles complex-1, subunit 3 |
| chr20_-_39599959 | 0.83 |
|
|
|
| chr15_+_14918610 | 0.83 |
ENSDART00000163066
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr20_+_27188382 | 0.83 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr23_-_42918086 | 0.83 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr15_+_1569674 | 0.83 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr21_+_6091802 | 0.83 |
ENSDART00000147716
|
si:dkey-93m18.3
|
si:dkey-93m18.3 |
| chr6_-_44163685 | 0.83 |
ENSDART00000035513
|
shq1
|
SHQ1, H/ACA ribonucleoprotein assembly factor |
| chr5_+_26925663 | 0.82 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr21_+_19040595 | 0.82 |
ENSDART00000145969
|
BX000991.1
|
ENSDARG00000092282 |
| chr18_+_20045544 | 0.80 |
|
|
|
| chr25_-_34596585 | 0.80 |
ENSDART00000171659
|
zgc:162611
|
zgc:162611 |
| chr14_+_25927045 | 0.79 |
ENSDART00000168673
ENSDART00000128971 |
gm2a
|
GM2 ganglioside activator |
| chr6_-_10492724 | 0.79 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr25_-_21409895 | 0.78 |
ENSDART00000010706
|
immp2l
|
inner mitochondrial membrane peptidase subunit 2 |
| chr22_+_23260413 | 0.78 |
|
|
|
| chr16_+_29756032 | 0.78 |
ENSDART00000103054
|
lysmd1
|
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr13_-_35781541 | 0.78 |
ENSDART00000147522
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr6_+_3313468 | 0.77 |
ENSDART00000011785
|
rad54l
|
RAD54-like (S. cerevisiae) |
| chr16_+_54861268 | 0.77 |
|
|
|
| chr17_-_45549323 | 0.76 |
|
|
|
| chr20_+_49380442 | 0.76 |
|
|
|
| chr17_+_24045786 | 0.76 |
ENSDART00000132755
|
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr23_-_42918055 | 0.76 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr16_+_38251717 | 0.75 |
ENSDART00000044971
|
myo1eb
|
myosin IE, b |
| chr19_+_47918720 | 0.75 |
ENSDART00000157886
|
zgc:114119
|
zgc:114119 |
| chr5_-_23741624 | 0.75 |
ENSDART00000042481
|
phf23a
|
PHD finger protein 23a |
| chr20_+_26005068 | 0.75 |
ENSDART00000016864
|
slc35f6
|
solute carrier family 35, member F6 |
| chr14_-_30564987 | 0.74 |
ENSDART00000173451
|
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr20_+_13245355 | 0.74 |
ENSDART00000034098
|
dtl
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr19_+_1148544 | 0.74 |
ENSDART00000166088
|
zgc:63863
|
zgc:63863 |
| chr2_+_36738864 | 0.72 |
ENSDART00000019063
|
pdcd10b
|
programmed cell death 10b |
| chr5_-_25133229 | 0.71 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr4_-_20092577 | 0.71 |
ENSDART00000164410
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr19_-_40111964 | 0.70 |
|
|
|
| chr2_+_5494310 | 0.70 |
ENSDART00000154167
|
sft2d3
|
SFT2 domain containing 3 |
| chr18_+_20045293 | 0.69 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr19_-_15324823 | 0.69 |
ENSDART00000169883
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr1_-_21188809 | 0.69 |
ENSDART00000169381
|
melk
|
maternal embryonic leucine zipper kinase |
| chr10_-_36848872 | 0.68 |
|
|
|
| chr3_+_19829535 | 0.68 |
|
|
|
| chr8_-_11164610 | 0.67 |
ENSDART00000147817
|
fam208b
|
family with sequence similarity 208, member B |
| chr7_-_56465369 | 0.66 |
ENSDART00000020967
|
csnk2a2a
|
casein kinase 2, alpha prime polypeptide a |
| chr5_-_37359835 | 0.66 |
ENSDART00000141059
|
mpzl2b
|
myelin protein zero-like 2b |
| chr3_-_34208507 | 0.66 |
ENSDART00000151634
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr19_+_1356915 | 0.66 |
ENSDART00000081779
|
zdhhc3b
|
zinc finger, DHHC-type containing 3b |
| chr8_+_8908955 | 0.65 |
ENSDART00000131215
|
slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member 2 |
| chr14_+_43357313 | 0.65 |
|
|
|
| chr18_-_40763266 | 0.65 |
ENSDART00000130397
|
akt2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr15_+_21776031 | 0.64 |
ENSDART00000136151
|
NKAPD1
|
zgc:162339 |
| chr22_-_17529086 | 0.64 |
|
|
|
| chr3_-_37429712 | 0.64 |
ENSDART00000161401
|
arf2a
|
ADP-ribosylation factor 2a |
| chr20_+_24047842 | 0.63 |
ENSDART00000128616
ENSDART00000144195 |
casp8ap2
|
caspase 8 associated protein 2 |
| chr5_+_26925957 | 0.63 |
|
|
|
| chr14_+_45774485 | 0.63 |
ENSDART00000146918
|
rnf44
|
ring finger protein 44 |
| chr8_+_3373146 | 0.63 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr16_+_25256694 | 0.62 |
ENSDART00000156416
|
wu:fe05a04
|
wu:fe05a04 |
| chr7_-_40307611 | 0.62 |
ENSDART00000173926
ENSDART00000010035 |
dnajb6b
|
DnaJ (Hsp40) homolog, subfamily B, member 6b |
| chr12_-_26445560 | 0.62 |
ENSDART00000138437
|
acsf2
|
acyl-CoA synthetase family member 2 |
| chr22_+_1153361 | 0.62 |
ENSDART00000159761
|
irf6
|
interferon regulatory factor 6 |
| chr3_+_19095975 | 0.62 |
ENSDART00000134514
|
smarca4a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4a |
| chr17_-_45549413 | 0.61 |
|
|
|
| chr5_-_23741680 | 0.61 |
ENSDART00000042481
|
phf23a
|
PHD finger protein 23a |
| chr23_-_42918166 | 0.60 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr24_-_1160886 | 0.60 |
|
|
|
| chr8_-_11164322 | 0.60 |
ENSDART00000147817
|
fam208b
|
family with sequence similarity 208, member B |
| chr12_+_28795843 | 0.59 |
ENSDART00000076342
|
rnf40
|
ring finger protein 40 |
| chr25_+_35159606 | 0.59 |
|
|
|
| chr3_+_25726394 | 0.59 |
ENSDART00000097679
|
zgc:171844
|
zgc:171844 |
| chr2_+_31855153 | 0.59 |
ENSDART00000066789
|
stard3nl
|
STARD3 N-terminal like |
| chr13_+_34563757 | 0.59 |
ENSDART00000133661
|
tasp1
|
taspase, threonine aspartase, 1 |
| chr16_+_54953101 | 0.59 |
|
|
|
| chr16_-_6909034 | 0.59 |
ENSDART00000149206
ENSDART00000149778 |
mbpb
|
myelin basic protein b |
| chr8_-_31407839 | 0.58 |
ENSDART00000098912
|
znf131
|
zinc finger protein 131 |
| chr8_-_11164125 | 0.58 |
ENSDART00000147817
|
fam208b
|
family with sequence similarity 208, member B |
| chr18_+_20044862 | 0.58 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr15_-_778461 | 0.58 |
ENSDART00000153874
|
si:dkey-7i4.19
|
si:dkey-7i4.19 |
| chr9_-_46609555 | 0.58 |
|
|
|
| chr20_-_16271738 | 0.57 |
ENSDART00000012476
|
coa7
|
cytochrome c oxidase assembly factor 7 |
| chr24_-_37178907 | 0.57 |
|
|
|
| chr2_-_30216240 | 0.57 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr18_-_40763107 | 0.57 |
ENSDART00000130397
|
akt2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr2_-_9898351 | 0.56 |
ENSDART00000165712
|
selt1a
|
selenoprotein T, 1a |
| chr15_+_38113014 | 0.56 |
|
|
|
| chr5_-_23741334 | 0.55 |
ENSDART00000042481
|
phf23a
|
PHD finger protein 23a |
| chr12_-_33457780 | 0.55 |
ENSDART00000161167
|
mbtd1
|
mbt domain containing 1 |
| chr2_-_37479961 | 0.55 |
ENSDART00000145896
|
si:dkey-57k2.7
|
si:dkey-57k2.7 |
| chr6_+_10098305 | 0.55 |
ENSDART00000151477
|
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr2_+_29468496 | 0.55 |
|
|
|
| chr2_-_30216703 | 0.55 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr14_+_32511519 | 0.54 |
ENSDART00000166351
|
nkrf
|
NFKB repressing factor |
| chr11_-_7786415 | 0.54 |
ENSDART00000154569
|
CR450813.1
|
ENSDARG00000097035 |
| chr17_-_24872401 | 0.54 |
ENSDART00000135569
|
gale
|
UDP-galactose-4-epimerase |
| chr15_-_5811394 | 0.53 |
ENSDART00000158522
|
hnrnpl2
|
heterogeneous nuclear ribonucleoprotein L2 |
| chr19_+_43277870 | 0.53 |
ENSDART00000135164
|
pdcd6ip
|
programmed cell death 6 interacting protein |
| chr7_+_26378093 | 0.53 |
ENSDART00000173823
ENSDART00000101053 |
tp53i11a
|
tumor protein p53 inducible protein 11a |
| chr6_-_14938420 | 0.52 |
ENSDART00000087797
|
tgfbrap1
|
transforming growth factor, beta receptor associated protein 1 |
| chr14_-_32467378 | 0.52 |
|
|
|
| chr16_+_43464824 | 0.52 |
ENSDART00000032778
|
rnf144b
|
ring finger protein 144B |
| chr12_-_28840748 | 0.52 |
|
|
|
| chr21_-_44748449 | 0.51 |
ENSDART00000026178
|
kif4
|
kinesin family member 4 |
| chr15_-_8880237 | 0.50 |
ENSDART00000008273
|
rab4b
|
RAB4B, member RAS oncogene family |
| chr25_+_35159867 | 0.50 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.8 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.7 | 2.1 | GO:0000493 | box H/ACA snoRNP assembly(GO:0000493) |
| 0.6 | 1.9 | GO:0003091 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.5 | 3.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.5 | 4.1 | GO:1901031 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.4 | 2.1 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.4 | 2.9 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.4 | 1.6 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.4 | 1.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.3 | 1.3 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.3 | 1.1 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.2 | 3.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 1.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.2 | 0.6 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.2 | 0.8 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.2 | 0.7 | GO:0072401 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.2 | 4.4 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.5 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 1.7 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.3 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.1 | 1.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.8 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 1.0 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.7 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 3.9 | GO:0035825 | reciprocal DNA recombination(GO:0035825) |
| 0.1 | 1.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 1.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 2.1 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 1.8 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 1.0 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 1.8 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 0.4 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.1 | 0.4 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 1.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 0.6 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.1 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.7 | GO:0032387 | negative regulation of intracellular transport(GO:0032387) |
| 0.1 | 1.7 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.1 | 0.4 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 0.9 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 1.1 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 0.9 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.2 | GO:0042665 | ectodermal cell fate commitment(GO:0001712) ectodermal cell fate specification(GO:0001715) ectodermal cell differentiation(GO:0010668) regulation of ectodermal cell fate specification(GO:0042665) regulation of ectoderm development(GO:2000383) |
| 0.1 | 0.3 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 1.6 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.1 | 0.2 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 4.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.7 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.6 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.3 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.9 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.3 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.0 | 0.8 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 0.2 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 1.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.8 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.8 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.2 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 1.2 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:2000480 | regulation of cAMP-dependent protein kinase activity(GO:2000479) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) amino sugar biosynthetic process(GO:0046349) |
| 0.0 | 0.2 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 1.2 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.9 | GO:0071600 | otic vesicle morphogenesis(GO:0071600) |
| 0.0 | 0.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 1.4 | GO:0007283 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 0.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.7 | GO:0061351 | neural precursor cell proliferation(GO:0061351) |
| 0.0 | 0.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:0060847 | endothelial cell fate specification(GO:0060847) blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.4 | GO:0048278 | membrane docking(GO:0022406) vesicle docking(GO:0048278) |
| 0.0 | 0.1 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0002144 | cytosolic tRNA wobble base thiouridylase complex(GO:0002144) |
| 0.5 | 4.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.4 | 1.2 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.4 | 1.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.3 | 1.2 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.2 | 0.7 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 1.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 1.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.3 | GO:0099631 | postsynaptic endocytic zone cytoplasmic component(GO:0099631) |
| 0.1 | 1.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.3 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.1 | 1.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.3 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 0.2 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 0.9 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 1.0 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.9 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 5.6 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 1.0 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.5 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 3.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.9 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 2.8 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.8 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.7 | GO:0016528 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 1.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.0 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.7 | GO:0030027 | lamellipodium(GO:0030027) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0047611 | acetylspermidine deacetylase activity(GO:0047611) |
| 0.6 | 1.9 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.5 | 4.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.4 | 5.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.4 | 2.0 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.4 | 1.6 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.3 | 0.9 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.3 | 3.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 4.4 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 1.8 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 1.3 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 1.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.8 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 1.4 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 1.7 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 1.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 1.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.0 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.1 | 2.0 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.1 | 0.5 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 2.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 0.2 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 0.7 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 2.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 3.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.7 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.7 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.9 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 1.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.0 | 5.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 1.8 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.6 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.0 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.5 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.3 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 2.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 3.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 0.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.2 | 2.2 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.2 | 1.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 1.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 2.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 5.1 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.1 | 0.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 3.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 1.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 0.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.2 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 3.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 1.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |