Project
DANIO-CODE
Navigation
Downloads
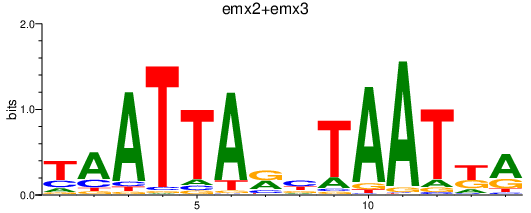
Results for emx2+emx3
Z-value: 1.67
Transcription factors associated with emx2+emx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
emx3
|
ENSDARG00000020417 | empty spiracles homeobox 3 |
|
emx2
|
ENSDARG00000039701 | empty spiracles homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| emx2 | dr10_dc_chr13_+_19191645_19191712 | -0.73 | 1.4e-03 | Click! |
| emx3 | dr10_dc_chr14_-_26078923_26078978 | -0.61 | 1.2e-02 | Click! |
Activity profile of emx2+emx3 motif
Sorted Z-values of emx2+emx3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of emx2+emx3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_10411573 | 6.92 |
ENSDART00000171232
|
ccdc106b
|
coiled-coil domain containing 106b |
| chr11_-_6442588 | 6.34 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr8_-_23759076 | 5.05 |
ENSDART00000145894
|
zgc:195245
|
zgc:195245 |
| chr1_-_18118467 | 4.61 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr19_-_20819477 | 4.58 |
ENSDART00000151356
|
dazl
|
deleted in azoospermia-like |
| chr19_-_20819101 | 4.40 |
ENSDART00000137590
|
dazl
|
deleted in azoospermia-like |
| chr13_+_29904945 | 4.15 |
ENSDART00000057525
|
dnajb12a
|
DnaJ (Hsp40) homolog, subfamily B, member 12a |
| chr18_-_40718244 | 3.96 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr21_+_6289363 | 3.90 |
ENSDART00000138600
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr22_+_17235696 | 3.57 |
ENSDART00000134798
|
tdrd5
|
tudor domain containing 5 |
| chr2_+_15432208 | 3.56 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr19_-_1571878 | 3.53 |
|
|
|
| chr19_-_10411518 | 3.52 |
ENSDART00000171232
|
ccdc106b
|
coiled-coil domain containing 106b |
| chr7_-_73016194 | 3.42 |
|
|
|
| chr2_+_15432130 | 3.34 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr4_+_9466175 | 3.17 |
ENSDART00000012659
|
zgc:55888
|
zgc:55888 |
| chr21_+_22598805 | 3.03 |
ENSDART00000142495
|
BX510940.1
|
ENSDARG00000089088 |
| chr5_+_37303599 | 3.03 |
ENSDART00000097754
ENSDART00000162470 |
tmprss4b
|
transmembrane protease, serine 4b |
| chr3_+_32285237 | 2.85 |
ENSDART00000157324
|
prrg2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr5_+_58936399 | 2.73 |
|
|
|
| chr19_-_18664720 | 2.72 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr15_+_35076414 | 2.68 |
ENSDART00000165210
|
zgc:66024
|
zgc:66024 |
| chr10_-_44713495 | 2.64 |
ENSDART00000076084
|
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr21_-_32027717 | 2.62 |
ENSDART00000131651
|
ENSDARG00000073961
|
ENSDARG00000073961 |
| chr3_+_12087549 | 2.60 |
|
|
|
| chr23_+_12225922 | 2.49 |
ENSDART00000136046
|
ppp1r3da
|
protein phosphatase 1, regulatory subunit 3Da |
| chr18_-_38787413 | 2.48 |
|
|
|
| chr13_+_849563 | 2.40 |
|
|
|
| chr21_+_31216418 | 2.39 |
ENSDART00000178521
|
asl
|
argininosuccinate lyase |
| chr4_-_3340315 | 2.38 |
ENSDART00000009076
|
pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr24_+_41991604 | 2.23 |
|
|
|
| chr17_-_1907622 | 2.21 |
ENSDART00000156489
|
xgb
|
x globin |
| chr1_+_18118735 | 2.20 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr19_-_10411878 | 2.12 |
ENSDART00000136653
|
ccdc106b
|
coiled-coil domain containing 106b |
| chr19_-_25565317 | 2.11 |
ENSDART00000175266
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr7_-_73613531 | 2.10 |
ENSDART00000128137
|
zgc:92594
|
zgc:92594 |
| chr7_+_66660882 | 2.07 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr21_+_13291305 | 2.04 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr21_-_44086464 | 2.03 |
ENSDART00000130833
|
FO704810.1
|
ENSDARG00000087492 |
| chr7_-_26226924 | 2.01 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr11_-_44756789 | 1.96 |
ENSDART00000161712
|
syngr2b
|
synaptogyrin 2b |
| chr22_+_2735606 | 1.90 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr14_-_6738260 | 1.89 |
ENSDART00000171792
|
rufy1
|
RUN and FYVE domain containing 1 |
| chr14_-_26127173 | 1.87 |
ENSDART00000088690
|
lman2
|
lectin, mannose-binding 2 |
| chr11_+_17849608 | 1.83 |
ENSDART00000020283
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr13_-_51743281 | 1.82 |
|
|
|
| KN149710v1_+_38638 | 1.79 |
|
|
|
| chr3_+_40022244 | 1.78 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr21_-_45323151 | 1.77 |
ENSDART00000151106
|
ENSDARG00000053404
|
ENSDARG00000053404 |
| chr25_-_20933295 | 1.77 |
ENSDART00000138985
ENSDART00000046298 |
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr21_-_2159827 | 1.76 |
ENSDART00000169897
|
zgc:171220
|
zgc:171220 |
| chr4_+_9466147 | 1.75 |
ENSDART00000012659
|
zgc:55888
|
zgc:55888 |
| chr3_+_33608070 | 1.75 |
|
|
|
| chr1_+_8468971 | 1.74 |
ENSDART00000081343
|
plk1
|
polo-like kinase 1 (Drosophila) |
| chr15_+_35089305 | 1.74 |
ENSDART00000156515
|
zgc:55621
|
zgc:55621 |
| chr12_+_20222000 | 1.72 |
ENSDART00000150020
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr21_-_2159785 | 1.72 |
ENSDART00000169897
|
zgc:171220
|
zgc:171220 |
| chr19_-_25564720 | 1.71 |
ENSDART00000148432
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr17_-_1907667 | 1.71 |
ENSDART00000156489
|
xgb
|
x globin |
| chr21_+_13291242 | 1.71 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr13_+_30441704 | 1.70 |
ENSDART00000010052
|
ppifa
|
peptidylprolyl isomerase Fa |
| chr11_-_6442490 | 1.69 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr10_+_27028355 | 1.66 |
ENSDART00000064111
|
faub
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed b |
| chr10_+_45108698 | 1.65 |
|
|
|
| chr2_+_1645259 | 1.65 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr15_-_44581550 | 1.57 |
|
|
|
| chr13_-_14950971 | 1.56 |
ENSDART00000157482
|
sfxn5a
|
sideroflexin 5a |
| chr19_-_5186692 | 1.55 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr2_-_17721810 | 1.52 |
ENSDART00000100201
ENSDART00000141188 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr19_-_20819057 | 1.51 |
ENSDART00000136826
|
dazl
|
deleted in azoospermia-like |
| chr5_+_6391432 | 1.51 |
ENSDART00000170564
ENSDART00000086666 |
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr21_+_13291347 | 1.48 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr20_-_22532817 | 1.48 |
ENSDART00000177772
|
BX276097.1
|
ENSDARG00000108228 |
| chr18_+_35153749 | 1.45 |
ENSDART00000151073
|
si:ch211-195m9.3
|
si:ch211-195m9.3 |
| chr2_+_42077559 | 1.45 |
ENSDART00000023208
|
ENSDARG00000014209
|
ENSDARG00000014209 |
| chr21_-_25358498 | 1.44 |
ENSDART00000167189
|
ppme1
|
protein phosphatase methylesterase 1 |
| chr8_+_36527653 | 1.43 |
ENSDART00000136418
ENSDART00000061378 |
sf3a1
|
splicing factor 3a, subunit 1 |
| chr2_-_31849916 | 1.43 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr17_-_9794176 | 1.41 |
|
|
|
| chr23_-_40642966 | 1.39 |
ENSDART00000153751
|
rnf146
|
ring finger protein 146 |
| chr12_-_3941450 | 1.38 |
ENSDART00000114857
|
ppp4cb
|
protein phosphatase 4, catalytic subunit b |
| chr15_+_34734212 | 1.38 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr11_-_44734403 | 1.37 |
ENSDART00000160656
|
afmid
|
arylformamidase |
| chr7_-_26226628 | 1.37 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr7_-_73613763 | 1.37 |
ENSDART00000128137
|
zgc:92594
|
zgc:92594 |
| chr3_+_37507517 | 1.37 |
ENSDART00000075039
|
gosr2
|
golgi SNAP receptor complex member 2 |
| chr2_-_37650547 | 1.36 |
ENSDART00000008302
|
insra
|
insulin receptor a |
| chr18_+_14715573 | 1.36 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr1_+_40894908 | 1.36 |
ENSDART00000111367
|
si:dkey-56e3.3
|
si:dkey-56e3.3 |
| chr6_-_7529384 | 1.35 |
ENSDART00000091836
ENSDART00000151697 |
ubn2a
|
ubinuclein 2a |
| chr19_-_20819028 | 1.35 |
ENSDART00000136826
|
dazl
|
deleted in azoospermia-like |
| chr6_-_46740359 | 1.33 |
ENSDART00000011970
|
zgc:66479
|
zgc:66479 |
| chr1_-_50395003 | 1.32 |
ENSDART00000035150
|
spast
|
spastin |
| chr10_-_44713414 | 1.32 |
ENSDART00000076084
|
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr4_-_5100307 | 1.31 |
ENSDART00000138817
|
tmem209
|
transmembrane protein 209 |
| chr23_+_45486958 | 1.31 |
ENSDART00000111126
|
psip1b
|
PC4 and SFRS1 interacting protein 1b |
| chr11_-_6442547 | 1.29 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr12_+_33257120 | 1.27 |
|
|
|
| chr25_+_20596613 | 1.27 |
ENSDART00000073648
|
kxd1
|
KxDL motif containing 1 |
| chr2_-_31850133 | 1.27 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr7_+_22042296 | 1.27 |
ENSDART00000123457
|
TMEM102
|
transmembrane protein 102 |
| chr23_-_17502717 | 1.25 |
|
|
|
| chr13_+_6091158 | 1.25 |
ENSDART00000025910
|
apmap
|
adipocyte plasma membrane associated protein |
| chr3_+_26212025 | 1.24 |
ENSDART00000163832
|
si:ch211-156b7.4
|
si:ch211-156b7.4 |
| chr12_-_33257026 | 1.24 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr7_+_13130111 | 1.23 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr13_+_40347099 | 1.23 |
|
|
|
| chr9_-_22014861 | 1.22 |
ENSDART00000101986
|
mrpl30
|
mitochondrial ribosomal protein L30 |
| chr1_+_35253862 | 1.22 |
ENSDART00000139636
|
zgc:152968
|
zgc:152968 |
| chr19_-_18664670 | 1.21 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr10_-_35313462 | 1.21 |
ENSDART00000139107
|
prr11
|
proline rich 11 |
| chr7_-_23623141 | 1.21 |
|
|
|
| chr25_-_13307171 | 1.21 |
ENSDART00000056723
|
gins3
|
GINS complex subunit 3 |
| chr23_-_40643077 | 1.19 |
ENSDART00000133356
|
rnf146
|
ring finger protein 146 |
| chr25_-_34596670 | 1.19 |
ENSDART00000171659
|
zgc:162611
|
zgc:162611 |
| chr16_-_30775394 | 1.19 |
|
|
|
| chr21_+_20439227 | 1.19 |
|
|
|
| chr9_+_31204464 | 1.18 |
|
|
|
| chr23_+_22858063 | 1.18 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr5_+_19429620 | 1.16 |
ENSDART00000088819
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr10_-_11364807 | 1.15 |
ENSDART00000039145
|
inip
|
ints3 and nabp interacting protein |
| chr5_+_31475897 | 1.15 |
ENSDART00000144510
|
zmat5
|
zinc finger, matrin-type 5 |
| chr24_+_29902165 | 1.14 |
ENSDART00000168422
ENSDART00000100092 ENSDART00000113304 |
frrs1b
|
ferric-chelate reductase 1b |
| chr15_-_16241412 | 1.14 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr14_-_33517822 | 1.13 |
ENSDART00000112268
|
ocrl
|
oculocerebrorenal syndrome of Lowe |
| chr23_-_25208472 | 1.13 |
ENSDART00000103989
ENSDART00000160278 |
idh3g
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr8_+_21405821 | 1.13 |
ENSDART00000142758
|
si:dkey-163f12.10
|
si:dkey-163f12.10 |
| chr9_-_22014768 | 1.13 |
ENSDART00000101986
|
mrpl30
|
mitochondrial ribosomal protein L30 |
| chr15_-_47351885 | 1.13 |
|
|
|
| chr3_+_39437497 | 1.13 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr20_+_484286 | 1.13 |
ENSDART00000026794
|
nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr4_-_4603294 | 1.12 |
ENSDART00000130601
|
CABZ01020840.1
|
ENSDARG00000090401 |
| chr1_+_30925353 | 1.12 |
ENSDART00000057880
|
poll
|
polymerase (DNA directed), lambda |
| chr21_+_10152870 | 1.10 |
|
|
|
| chr3_+_37673971 | 1.10 |
|
|
|
| chr22_+_699477 | 1.09 |
ENSDART00000017305
|
znf76
|
zinc finger protein 76 |
| chr24_-_32668102 | 1.09 |
ENSDART00000066590
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr5_-_20252709 | 1.09 |
|
|
|
| chr18_+_15695287 | 1.09 |
|
|
|
| chr5_-_11309164 | 1.08 |
ENSDART00000132564
|
nf2a
|
neurofibromin 2a (merlin) |
| chr12_-_308764 | 1.08 |
ENSDART00000066579
ENSDART00000171396 |
pts
|
6-pyruvoyltetrahydropterin synthase |
| chr24_+_36316070 | 1.08 |
ENSDART00000155260
|
rbbp8
|
retinoblastoma binding protein 8 |
| chr14_+_51515192 | 1.05 |
|
|
|
| chr12_+_47828020 | 1.04 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr3_+_37507573 | 1.04 |
ENSDART00000075039
|
gosr2
|
golgi SNAP receptor complex member 2 |
| chr10_+_35313772 | 1.03 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr22_+_35299551 | 1.01 |
ENSDART00000165353
|
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr7_+_13130177 | 1.01 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr19_-_2928582 | 1.00 |
ENSDART00000109130
|
recql4
|
RecQ helicase-like 4 |
| chr23_+_26099718 | 1.00 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr11_-_25224150 | 1.00 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr8_+_42910733 | 1.00 |
ENSDART00000021715
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr12_+_47097965 | 0.99 |
ENSDART00000084714
|
zranb1a
|
zinc finger, RAN-binding domain containing 1a |
| chr11_-_36425176 | 0.99 |
ENSDART00000156166
|
CR457444.1
|
ENSDARG00000096865 |
| chr8_-_22252628 | 0.99 |
ENSDART00000121513
|
nphp4
|
nephronophthisis 4 |
| chr23_+_29956616 | 0.98 |
ENSDART00000148706
|
aurkaip1
|
aurora kinase A interacting protein 1 |
| chr6_-_52725402 | 0.97 |
ENSDART00000033949
|
oser1
|
oxidative stress responsive serine-rich 1 |
| chr8_-_44247277 | 0.97 |
|
|
|
| chr17_+_48914379 | 0.97 |
|
|
|
| chr11_-_10867083 | 0.96 |
ENSDART00000091901
|
psmd14
|
proteasome 26S subunit, non-ATPase 14 |
| chr23_-_40643024 | 0.96 |
ENSDART00000153751
|
rnf146
|
ring finger protein 146 |
| chr22_-_17571930 | 0.96 |
|
|
|
| chr23_-_27647386 | 0.95 |
|
|
|
| chr8_+_53202445 | 0.95 |
|
|
|
| chr14_+_37317006 | 0.93 |
|
|
|
| chr3_-_48458042 | 0.93 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr6_+_3556296 | 0.93 |
ENSDART00000041627
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr9_+_24084807 | 0.92 |
ENSDART00000131953
|
cops8
|
COP9 signalosome subunit 8 |
| chr12_+_27151693 | 0.92 |
ENSDART00000066269
|
arl4d
|
ADP-ribosylation factor-like 4D |
| chr25_+_3091855 | 0.92 |
ENSDART00000104859
|
rccd1
|
RCC1 domain containing 1 |
| chr7_+_8834029 | 0.92 |
ENSDART00000173250
|
pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr21_-_25765319 | 0.91 |
ENSDART00000101219
|
mettl27
|
methyltransferase like 27 |
| chr13_+_35402766 | 0.91 |
ENSDART00000163368
ENSDART00000075414 ENSDART00000112947 |
wdr27
|
WD repeat domain 27 |
| chr11_-_8116290 | 0.90 |
ENSDART00000165569
|
ttll7
|
tubulin tyrosine ligase-like family, member 7 |
| chr10_+_43953171 | 0.88 |
|
|
|
| chr21_-_1618494 | 0.87 |
ENSDART00000124904
|
zgc:152948
|
zgc:152948 |
| chr24_-_18732618 | 0.87 |
ENSDART00000092783
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr22_+_17235538 | 0.86 |
ENSDART00000134798
|
tdrd5
|
tudor domain containing 5 |
| chr9_+_26029780 | 0.86 |
ENSDART00000127135
|
gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr8_-_53593850 | 0.86 |
|
|
|
| chr5_+_58936508 | 0.85 |
|
|
|
| chr20_+_25687135 | 0.84 |
ENSDART00000063122
|
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr24_+_16402587 | 0.84 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr7_-_48394268 | 0.83 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr5_+_19429500 | 0.83 |
ENSDART00000168868
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr8_-_31706783 | 0.82 |
ENSDART00000061832
|
si:dkey-46a10.3
|
si:dkey-46a10.3 |
| chr23_-_29825098 | 0.82 |
ENSDART00000056865
|
ctnnbip1
|
catenin, beta interacting protein 1 |
| chr4_+_13413303 | 0.82 |
ENSDART00000003694
|
cand1
|
cullin-associated and neddylation-dissociated 1 |
| chr11_-_10472636 | 0.82 |
ENSDART00000081827
|
ect2
|
epithelial cell transforming 2 |
| chr7_+_51499409 | 0.81 |
ENSDART00000007767
|
got2b
|
glutamic-oxaloacetic transaminase 2b, mitochondrial |
| chr14_-_10081301 | 0.81 |
ENSDART00000165101
|
commd5
|
COMM domain containing 5 |
| chr19_-_43018998 | 0.81 |
ENSDART00000102583
|
sytl1
|
synaptotagmin-like 1 |
| chr14_+_23420053 | 0.81 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr8_-_39850553 | 0.80 |
ENSDART00000131372
|
mlec
|
malectin |
| chr2_-_27892824 | 0.80 |
|
|
|
| chr3_-_43393284 | 0.79 |
ENSDART00000157902
|
kdm8
|
lysine (K)-specific demethylase 8 |
| chr16_+_30393572 | 0.79 |
|
|
|
| chr23_+_22954156 | 0.79 |
|
|
|
| chr12_+_20000577 | 0.79 |
ENSDART00000161505
|
mgrn1a
|
mahogunin, ring finger 1a |
| chr4_+_8669695 | 0.78 |
ENSDART00000168768
|
adipor2
|
adiponectin receptor 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 11.8 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 1.0 | 3.9 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.5 | 2.6 | GO:0099541 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.5 | 1.5 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.5 | 5.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.5 | 1.9 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.4 | 4.4 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.4 | 1.7 | GO:1903726 | negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.4 | 1.6 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.3 | 1.0 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.3 | 1.3 | GO:0043243 | positive regulation of protein complex disassembly(GO:0043243) |
| 0.3 | 1.0 | GO:0071947 | protein K29-linked deubiquitination(GO:0035523) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.3 | 1.1 | GO:0071871 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.3 | 1.1 | GO:0015809 | arginine transport(GO:0015809) |
| 0.3 | 2.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.3 | 1.1 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.3 | 2.4 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.3 | 1.6 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.3 | 0.8 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.3 | 2.0 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.2 | 1.0 | GO:0015882 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.2 | 4.6 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.2 | 0.8 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 1.0 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.2 | 1.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 4.0 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.2 | 1.2 | GO:1902292 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.2 | 1.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.2 | 0.5 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 0.8 | GO:1903513 | endoplasmic reticulum to cytosol transport(GO:1903513) |
| 0.1 | 1.0 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.1 | 6.1 | GO:0007599 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.1 | 2.5 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 3.0 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.1 | 0.5 | GO:0031645 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.1 | 1.6 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.9 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.8 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.1 | 3.5 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 1.7 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.8 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.7 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 1.0 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 1.0 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 0.3 | GO:0046950 | cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.1 | 0.6 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.1 | 0.9 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.7 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 | 1.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 3.4 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.1 | 1.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.8 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 0.3 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 1.4 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.1 | 0.4 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.1 | 0.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 1.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.5 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 1.6 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.1 | 0.4 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 0.2 | GO:1903010 | positive regulation of cilium assembly(GO:0045724) regulation of bone development(GO:1903010) |
| 0.1 | 1.7 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.1 | 1.1 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 2.0 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.3 | GO:0030817 | regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 1.4 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.9 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0097354 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.6 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.4 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.5 | GO:0009620 | response to yeast(GO:0001878) response to fungus(GO:0009620) |
| 0.0 | 0.7 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.0 | 0.1 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 1.1 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.0 | 0.8 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.8 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.1 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0046498 | S-adenosylmethionine cycle(GO:0033353) S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 1.2 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.7 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.5 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 1.0 | GO:0000723 | telomere maintenance(GO:0000723) telomere organization(GO:0032200) |
| 0.0 | 0.2 | GO:0006895 | phagolysosome assembly(GO:0001845) Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.6 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.5 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.4 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.2 | GO:0035794 | positive regulation of mitochondrial membrane permeability(GO:0035794) mitochondrial outer membrane permeabilization(GO:0097345) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.0 | 0.2 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.3 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.3 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.2 | GO:0098781 | ncRNA transcription(GO:0098781) |
| 0.0 | 0.6 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.6 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of chemotaxis(GO:0050922) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.3 | GO:0007520 | myoblast fusion(GO:0007520) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.5 | 2.7 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.3 | 1.0 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.2 | 1.7 | GO:0031838 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 1.2 | GO:0000811 | GINS complex(GO:0000811) |
| 0.2 | 1.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 1.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 1.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.4 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 2.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 1.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.3 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 1.1 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 1.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.6 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.4 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 0.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 2.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 3.6 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 1.0 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 1.5 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.3 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 1.9 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 1.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.5 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.0 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.3 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 2.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 5.5 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 1.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 2.2 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 0.9 | GO:0022627 | cytosolic ribosome(GO:0022626) cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.1 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 2.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 14.0 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 1.0 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.0 | 0.4 | GO:0005876 | spindle microtubule(GO:0005876) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 11.8 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.9 | 3.5 | GO:0072572 | poly-ADP-D-ribose binding(GO:0072572) |
| 0.7 | 3.9 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.6 | 2.4 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.5 | 1.6 | GO:0000403 | Y-form DNA binding(GO:0000403) double-strand/single-strand DNA junction binding(GO:0000406) flap-structured DNA binding(GO:0070336) |
| 0.5 | 1.5 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.5 | 1.9 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.4 | 2.6 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.4 | 1.3 | GO:0016843 | amine-lyase activity(GO:0016843) strictosidine synthase activity(GO:0016844) |
| 0.4 | 1.1 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.3 | 1.8 | GO:0016972 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.3 | 1.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.3 | 1.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.3 | 1.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.3 | 4.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.2 | 1.7 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.2 | 0.7 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.2 | 0.7 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.2 | 1.5 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 2.5 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.2 | 1.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 1.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 6.1 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 1.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.4 | GO:0003874 | 6-pyruvoyltetrahydropterin synthase activity(GO:0003874) |
| 0.1 | 1.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.0 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.1 | 3.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.7 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 1.7 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.5 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 1.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.8 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 1.1 | GO:0034595 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.1 | 0.3 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.1 | 0.5 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.8 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 1.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.6 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 2.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.1 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 2.0 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.8 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.3 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 1.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 5.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.8 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 3.0 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.1 | GO:0016801 | hydrolase activity, acting on ether bonds(GO:0016801) |
| 0.0 | 0.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 2.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 1.4 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 2.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.6 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.0 | 0.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.9 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 2.2 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.3 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 2.7 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 4.9 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 3.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 2.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 0.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 2.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 1.6 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 1.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.1 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 2.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 1.9 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 2.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 0.7 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.3 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.1 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.6 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.7 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |