Project
DANIO-CODE
Navigation
Downloads
Results for en1a
Z-value: 0.67
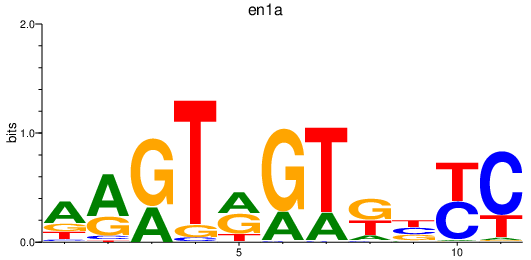
Transcription factors associated with en1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
en1a
|
ENSDARG00000014321 | engrailed homeobox 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| en1a | dr10_dc_chr9_-_785569_785606 | -0.77 | 4.6e-04 | Click! |
Activity profile of en1a motif
Sorted Z-values of en1a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of en1a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_-_36621626 | 1.90 |
ENSDART00000128108
|
urahb
|
urate (5-hydroxyiso-) hydrolase b |
| chr3_-_39316317 | 1.77 |
|
|
|
| chr1_+_41762057 | 1.55 |
ENSDART00000137609
|
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr2_-_2373601 | 1.42 |
ENSDART00000113774
|
si:ch211-188c16.1
|
si:ch211-188c16.1 |
| chr23_-_33812361 | 1.34 |
ENSDART00000136386
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr13_+_8508786 | 1.25 |
ENSDART00000059321
|
epcam
|
epithelial cell adhesion molecule |
| chr9_-_11616001 | 1.24 |
ENSDART00000030995
|
umps
|
uridine monophosphate synthetase |
| chr24_-_9865837 | 1.20 |
ENSDART00000106244
|
zgc:171750
|
zgc:171750 |
| chr1_+_21964075 | 1.17 |
ENSDART00000161874
|
anxa5b
|
annexin A5b |
| chr17_-_1524813 | 1.12 |
ENSDART00000112803
|
wdr20a
|
WD repeat domain 20a |
| chr6_+_39186673 | 1.11 |
ENSDART00000156187
|
tac3b
|
tachykinin 3b |
| chr10_-_25448769 | 1.10 |
ENSDART00000140023
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr23_+_6652454 | 1.10 |
ENSDART00000081763
|
rbm38
|
RNA binding motif protein 38 |
| chr18_-_12889296 | 1.09 |
ENSDART00000130343
|
parp12a
|
poly (ADP-ribose) polymerase family, member 12a |
| chr23_+_19663746 | 1.06 |
ENSDART00000170149
|
slmapb
|
sarcolemma associated protein b |
| chr17_-_31387062 | 1.01 |
|
|
|
| chr3_-_27515161 | 0.97 |
ENSDART00000151027
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr21_-_7322856 | 0.91 |
ENSDART00000151543
ENSDART00000114982 |
f2rl1.2
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 2 |
| chr5_-_32433490 | 0.89 |
|
|
|
| chr16_+_47283253 | 0.89 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr16_+_41065840 | 0.88 |
ENSDART00000124543
|
dek
|
DEK proto-oncogene |
| chr24_-_16835352 | 0.87 |
ENSDART00000005331
ENSDART00000128446 |
klhl15
|
kelch-like family member 15 |
| chr17_-_18777398 | 0.86 |
ENSDART00000045991
|
vrk1
|
vaccinia related kinase 1 |
| chr5_+_58883315 | 0.85 |
|
|
|
| chr2_+_36623514 | 0.85 |
ENSDART00000098415
|
ing5b
|
inhibitor of growth family, member 5b |
| chr14_-_47210912 | 0.81 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr12_-_10327938 | 0.79 |
ENSDART00000007335
|
ndc80
|
NDC80 kinetochore complex component |
| chr5_-_41396159 | 0.79 |
ENSDART00000143573
|
ncor1
|
nuclear receptor corepressor 1 |
| chr19_-_3547196 | 0.75 |
ENSDART00000166107
|
hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr23_-_17055508 | 0.75 |
ENSDART00000125472
|
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr18_+_19467527 | 0.74 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr7_-_18460264 | 0.73 |
ENSDART00000173905
|
trmt10a
|
tRNA methyltransferase 10A |
| chr5_-_57053687 | 0.72 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr10_-_22180856 | 0.72 |
ENSDART00000006173
|
cldn7b
|
claudin 7b |
| chr7_-_20330881 | 0.71 |
ENSDART00000169750
|
si:dkey-19b23.11
|
si:dkey-19b23.11 |
| chr18_-_12889369 | 0.69 |
ENSDART00000130343
|
parp12a
|
poly (ADP-ribose) polymerase family, member 12a |
| chr3_-_42949449 | 0.68 |
ENSDART00000161127
|
axin1
|
axin 1 |
| chr7_+_26730804 | 0.68 |
|
|
|
| chr22_+_818795 | 0.68 |
ENSDART00000003325
|
dennd2db
|
DENN/MADD domain containing 2Db |
| chr13_+_8509344 | 0.68 |
ENSDART00000140489
|
epcam
|
epithelial cell adhesion molecule |
| chr20_+_52021000 | 0.68 |
ENSDART00000074330
|
taf1a
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A |
| chr14_+_23420053 | 0.67 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr10_-_22180935 | 0.67 |
ENSDART00000006173
|
cldn7b
|
claudin 7b |
| chr12_-_20494169 | 0.67 |
ENSDART00000105362
|
snx11
|
sorting nexin 11 |
| chr3_-_26060787 | 0.66 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr6_+_38898605 | 0.65 |
ENSDART00000029930
|
slc48a1b
|
solute carrier family 48 (heme transporter), member 1b |
| chr3_-_21006698 | 0.63 |
ENSDART00000104051
|
cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr25_+_34555948 | 0.63 |
ENSDART00000111706
|
si:dkey-108k21.14
|
si:dkey-108k21.14 |
| chr3_-_42949616 | 0.63 |
ENSDART00000161127
|
axin1
|
axin 1 |
| chr21_-_31215463 | 0.62 |
ENSDART00000170507
|
crcp
|
calcitonin gene-related peptide-receptor component protein |
| chr21_-_21787648 | 0.62 |
ENSDART00000030984
|
wdr73
|
WD repeat domain 73 |
| chr19_+_12730217 | 0.62 |
ENSDART00000088917
|
rnmt
|
RNA (guanine-7-) methyltransferase |
| chr19_+_43316957 | 0.62 |
ENSDART00000151298
|
arpp21
|
cAMP-regulated phosphoprotein, 21 |
| chr10_-_25448712 | 0.60 |
ENSDART00000140023
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr12_+_9811557 | 0.60 |
ENSDART00000110458
|
fam117ab
|
family with sequence similarity 117, member Ab |
| chr7_+_28824986 | 0.59 |
ENSDART00000052349
|
ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr17_+_13292569 | 0.59 |
ENSDART00000177104
|
BX005308.2
|
ENSDARG00000108498 |
| chr2_+_37855019 | 0.58 |
ENSDART00000113337
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr12_-_26445609 | 0.58 |
ENSDART00000132737
ENSDART00000163931 |
acsf2
|
acyl-CoA synthetase family member 2 |
| chr2_-_26934833 | 0.58 |
ENSDART00000025120
|
ndc1
|
NDC1 transmembrane nucleoporin |
| chr3_-_42949526 | 0.57 |
ENSDART00000161127
|
axin1
|
axin 1 |
| chr5_+_60782855 | 0.56 |
ENSDART00000074073
|
alkbh4
|
alkB homolog 4, lysine demthylase |
| chr2_+_38042821 | 0.55 |
ENSDART00000134211
ENSDART00000144868 |
hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr22_+_18840143 | 0.54 |
|
|
|
| chr23_-_17055376 | 0.54 |
ENSDART00000125472
|
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr18_+_12853364 | 0.54 |
|
|
|
| chr6_+_38898811 | 0.54 |
ENSDART00000131347
|
slc48a1b
|
solute carrier family 48 (heme transporter), member 1b |
| chr21_-_21787540 | 0.53 |
ENSDART00000030984
|
wdr73
|
WD repeat domain 73 |
| chr16_-_10946422 | 0.52 |
ENSDART00000036891
|
rabac1
|
Rab acceptor 1 (prenylated) |
| chr17_+_5776013 | 0.52 |
ENSDART00000168326
|
znf513b
|
zinc finger protein 513b |
| chr18_-_16927749 | 0.52 |
ENSDART00000100105
ENSDART00000163400 |
swap70b
|
SWAP switching B-cell complex 70b subunit 70b |
| chr21_-_9291682 | 0.51 |
ENSDART00000169275
|
sdad1
|
SDA1 domain containing 1 |
| chr7_-_46940193 | 0.50 |
ENSDART00000161558
|
BX005073.2
|
ENSDARG00000099964 |
| chr3_-_42949479 | 0.50 |
ENSDART00000161127
|
axin1
|
axin 1 |
| chr19_-_3849408 | 0.50 |
ENSDART00000165917
|
btr18
|
bloodthirsty-related gene family, member 18 |
| chr17_-_1524751 | 0.49 |
ENSDART00000112803
|
wdr20a
|
WD repeat domain 20a |
| chr22_-_24957034 | 0.49 |
ENSDART00000142147
|
dnal4b
|
dynein, axonemal, light chain 4b |
| chr2_-_22875246 | 0.49 |
ENSDART00000159641
|
znf644a
|
zinc finger protein 644a |
| chr16_+_54906490 | 0.48 |
ENSDART00000126646
|
ENSDARG00000091079
|
ENSDARG00000091079 |
| chr25_+_18997440 | 0.47 |
ENSDART00000155569
|
si:dkeyp-19h3.8
|
si:dkeyp-19h3.8 |
| chr21_-_9291452 | 0.47 |
ENSDART00000169275
|
sdad1
|
SDA1 domain containing 1 |
| chr10_+_15382807 | 0.47 |
ENSDART00000168909
|
trappc13
|
trafficking protein particle complex 13 |
| chr17_-_1524721 | 0.46 |
ENSDART00000112803
|
wdr20a
|
WD repeat domain 20a |
| chr24_+_10162691 | 0.45 |
ENSDART00000066794
ENSDART00000112455 |
otulina
otulina
|
OTU deubiquitinase with linear linkage specificity a OTU deubiquitinase with linear linkage specificity a |
| chr12_-_10343382 | 0.45 |
ENSDART00000152788
|
mki67
|
marker of proliferation Ki-67 |
| chr2_-_22836607 | 0.44 |
ENSDART00000168022
|
gtf2b
|
general transcription factor IIB |
| chr3_-_29779725 | 0.44 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr2_+_45119713 | 0.43 |
ENSDART00000075146
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr7_+_28825033 | 0.43 |
ENSDART00000052349
|
ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr23_-_17055458 | 0.42 |
ENSDART00000125472
|
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr20_-_37910887 | 0.41 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr20_-_44562847 | 0.41 |
ENSDART00000085442
|
mut
|
methylmalonyl CoA mutase |
| chr24_+_35296732 | 0.41 |
ENSDART00000172652
|
pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr9_-_12623750 | 0.41 |
ENSDART00000142024
|
tra2b
|
transformer 2 beta homolog |
| chr6_-_53391354 | 0.40 |
ENSDART00000155483
|
si:ch211-161c3.6
|
si:ch211-161c3.6 |
| chr4_+_5823740 | 0.40 |
ENSDART00000121743
|
si:ch73-352p4.5
|
si:ch73-352p4.5 |
| chr3_-_29779598 | 0.40 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr24_-_8585312 | 0.38 |
|
|
|
| chr12_+_23939869 | 0.38 |
ENSDART00000021298
|
asb3
|
ankyrin repeat and SOCS box containing 3 |
| chr23_-_21608362 | 0.38 |
ENSDART00000010647
|
rcc2
|
regulator of chromosome condensation 2 |
| chr20_-_34244538 | 0.37 |
ENSDART00000152963
|
BX248238.2
|
ENSDARG00000096789 |
| chr17_-_9794176 | 0.36 |
|
|
|
| chr3_-_33362384 | 0.36 |
ENSDART00000020342
|
sgsm3
|
small G protein signaling modulator 3 |
| chr12_-_25006119 | 0.36 |
ENSDART00000158036
|
cript
|
cysteine-rich PDZ-binding protein |
| chr23_+_116397 | 0.36 |
|
|
|
| chr21_-_10962154 | 0.35 |
ENSDART00000084061
|
nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like |
| chr4_+_25923205 | 0.35 |
ENSDART00000136927
|
vetz
|
vezatin, adherens junctions transmembrane protein |
| chr18_+_41242278 | 0.34 |
ENSDART00000138552
ENSDART00000145863 |
trip12
|
thyroid hormone receptor interactor 12 |
| chr21_-_31215400 | 0.34 |
ENSDART00000121946
|
crcp
|
calcitonin gene-related peptide-receptor component protein |
| chr9_+_23192282 | 0.34 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr4_+_10605257 | 0.34 |
ENSDART00000122636
ENSDART00000037140 |
ing3
|
inhibitor of growth family, member 3 |
| chr20_-_26517489 | 0.34 |
|
|
|
| chr4_+_13490875 | 0.33 |
ENSDART00000178376
|
BX465210.1
|
ENSDARG00000107553 |
| chr15_+_14918610 | 0.33 |
ENSDART00000163066
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr23_-_25853364 | 0.33 |
ENSDART00000110670
|
ENSDARG00000078574
|
ENSDARG00000078574 |
| chr18_-_7138414 | 0.31 |
ENSDART00000003748
|
cfap161
|
cilia and flagella associated protein 161 |
| chr18_-_510340 | 0.30 |
ENSDART00000164374
|
si:ch211-79l17.1
|
si:ch211-79l17.1 |
| chr5_+_23559435 | 0.29 |
ENSDART00000142268
|
gps2
|
G protein pathway suppressor 2 |
| chr3_-_29779552 | 0.29 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr10_+_20435169 | 0.29 |
ENSDART00000160803
|
r3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr14_-_43204831 | 0.28 |
ENSDART00000122339
|
gar1
|
GAR1 homolog, ribonucleoprotein |
| chr23_+_22408825 | 0.28 |
ENSDART00000147696
|
rap1gap
|
RAP1 GTPase activating protein |
| chr2_-_7772932 | 0.28 |
ENSDART00000160053
|
ripk2
|
receptor-interacting serine-threonine kinase 2 |
| chr25_-_28920272 | 0.28 |
ENSDART00000166889
|
nptna
|
neuroplastin a |
| chr11_-_35494404 | 0.27 |
ENSDART00000103076
|
arl8bb
|
ADP-ribosylation factor-like 8Bb |
| chr1_-_29764204 | 0.27 |
ENSDART00000144297
ENSDART00000164204 |
ubac2
|
UBA domain containing 2 |
| chr3_+_32802899 | 0.27 |
|
|
|
| chr22_-_10410949 | 0.27 |
ENSDART00000111962
ENSDART00000064809 |
nol8
|
nucleolar protein 8 |
| chr22_-_28695519 | 0.27 |
ENSDART00000005883
ENSDART00000007791 |
jagn1b
|
jagunal homolog 1b |
| chr16_+_19223553 | 0.27 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr23_-_26193648 | 0.27 |
ENSDART00000164032
|
rca2.1
|
regulator of complement activation group 2 gene 1 |
| chr14_-_40482957 | 0.27 |
ENSDART00000173236
|
ef1
|
E74-like factor 1 (ets domain transcription factor) |
| chr4_-_5822882 | 0.26 |
ENSDART00000008898
|
foxm1
|
forkhead box M1 |
| chr10_+_44846866 | 0.26 |
|
|
|
| chr1_-_37452112 | 0.26 |
ENSDART00000142811
|
hmgb2a
|
high mobility group box 2a |
| chr15_+_35185174 | 0.26 |
ENSDART00000086954
ENSDART00000161594 |
sesn3
|
sestrin 3 |
| chr5_+_53633496 | 0.26 |
ENSDART00000166050
|
rangrf
|
RAN guanine nucleotide release factor |
| chr19_-_307085 | 0.26 |
ENSDART00000145916
|
lingo4a
|
leucine rich repeat and Ig domain containing 4a |
| chr6_-_22910576 | 0.26 |
ENSDART00000164114
|
grb2a
|
growth factor receptor-bound protein 2a |
| chr11_-_6870810 | 0.24 |
ENSDART00000007204
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr5_-_68943821 | 0.24 |
ENSDART00000158956
|
mob1a
|
MOB kinase activator 1A |
| chr5_+_23559189 | 0.23 |
ENSDART00000142268
|
gps2
|
G protein pathway suppressor 2 |
| chr24_-_16835253 | 0.23 |
ENSDART00000005331
ENSDART00000128446 |
klhl15
|
kelch-like family member 15 |
| chr15_+_17505794 | 0.21 |
ENSDART00000153729
|
snx19b
|
sorting nexin 19b |
| chr14_-_7000549 | 0.21 |
ENSDART00000158914
|
si:ch211-51f19.1
|
si:ch211-51f19.1 |
| chr24_+_35296642 | 0.21 |
ENSDART00000172652
|
pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr17_-_5918853 | 0.21 |
ENSDART00000058890
ENSDART00000171084 |
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr13_-_34732375 | 0.21 |
|
|
|
| chr8_+_36951655 | 0.20 |
|
|
|
| chr6_+_32339159 | 0.20 |
ENSDART00000042134
|
dock7
|
dedicator of cytokinesis 7 |
| chr1_+_40763274 | 0.19 |
ENSDART00000135280
|
paip2b
|
poly(A) binding protein interacting protein 2B |
| chr4_-_29167253 | 0.18 |
ENSDART00000160579
|
CR388132.2
|
ENSDARG00000101782 |
| chr7_-_6334242 | 0.18 |
ENSDART00000081359
|
zgc:110425
|
zgc:110425 |
| chr11_-_6870835 | 0.18 |
ENSDART00000007204
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr7_+_49391403 | 0.17 |
ENSDART00000131210
|
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr7_+_32626787 | 0.17 |
ENSDART00000140800
ENSDART00000075263 ENSDART00000137956 |
tssc4
|
tumor suppressing subtransferable candidate 4 |
| chr23_-_17055590 | 0.16 |
ENSDART00000143180
|
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr1_+_40762892 | 0.16 |
ENSDART00000010211
|
paip2b
|
poly(A) binding protein interacting protein 2B |
| chr15_-_26998580 | 0.16 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| chr3_+_60035494 | 0.15 |
ENSDART00000166982
ENSDART00000166837 |
sec14l1
|
SEC14-like lipid binding 1 |
| chr5_-_68943600 | 0.15 |
ENSDART00000159851
|
mob1a
|
MOB kinase activator 1A |
| chr2_-_4277605 | 0.15 |
|
|
|
| chr20_-_26517630 | 0.15 |
|
|
|
| chr10_+_16207230 | 0.15 |
ENSDART00000065045
|
prrc1
|
proline-rich coiled-coil 1 |
| chr16_-_10946309 | 0.15 |
ENSDART00000049148
|
rabac1
|
Rab acceptor 1 (prenylated) |
| chr25_-_26309649 | 0.14 |
ENSDART00000156364
|
usp3
|
ubiquitin specific peptidase 3 |
| chr1_-_37452393 | 0.13 |
ENSDART00000047159
|
hmgb2a
|
high mobility group box 2a |
| chr1_-_54453712 | 0.13 |
ENSDART00000074083
|
rab1aa
|
RAB1A, member RAS oncogene family a |
| chr14_-_30150071 | 0.13 |
ENSDART00000173107
|
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr7_-_33791033 | 0.12 |
ENSDART00000052404
|
map2k5
|
mitogen-activated protein kinase kinase 5 |
| chr6_-_19556328 | 0.11 |
ENSDART00000074256
|
ppp1r12c
|
protein phosphatase 1, regulatory subunit 12C |
| chr23_-_25853331 | 0.11 |
ENSDART00000110670
|
ENSDARG00000078574
|
ENSDARG00000078574 |
| chr1_-_37451594 | 0.11 |
ENSDART00000142811
|
hmgb2a
|
high mobility group box 2a |
| chr7_-_6231350 | 0.10 |
ENSDART00000159524
|
CU457819.1
|
Histone H3.2 |
| chr6_-_8156520 | 0.08 |
ENSDART00000081561
|
ilf3a
|
interleukin enhancer binding factor 3a |
| chr9_+_12473243 | 0.08 |
ENSDART00000132709
|
tmem41aa
|
transmembrane protein 41aa |
| chr25_-_26309873 | 0.08 |
ENSDART00000155698
|
usp3
|
ubiquitin specific peptidase 3 |
| chr16_+_23468242 | 0.08 |
ENSDART00000103220
|
krtcap2
|
keratinocyte associated protein 2 |
| chr11_-_26463925 | 0.08 |
ENSDART00000083010
|
acad9
|
acyl-CoA dehydrogenase family, member 9 |
| chr9_-_21042485 | 0.07 |
ENSDART00000028247
|
gdap2
|
ganglioside induced differentiation associated protein 2 |
| chr25_+_18997277 | 0.07 |
ENSDART00000154066
|
isg20
|
interferon stimulated exonuclease gene |
| chr3_-_23466232 | 0.07 |
ENSDART00000156897
|
ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr18_-_29920047 | 0.07 |
ENSDART00000138533
|
cmc2
|
C-x(9)-C motif containing 2 |
| chr12_-_3803808 | 0.06 |
ENSDART00000160967
|
taok2b
|
TAO kinase 2b |
| chr21_+_34780385 | 0.05 |
ENSDART00000003962
|
wdr55
|
WD repeat domain 55 |
| chr9_-_30744878 | 0.05 |
ENSDART00000079222
|
chaf1b
|
chromatin assembly factor 1, subunit B |
| chr14_+_10984191 | 0.05 |
ENSDART00000113567
|
rlim
|
ring finger protein, LIM domain interacting |
| chr3_-_42949225 | 0.05 |
ENSDART00000161127
|
axin1
|
axin 1 |
| chr20_-_33887129 | 0.05 |
ENSDART00000020183
|
fam102bb
|
family with sequence similarity 102, member B, b |
| chr15_+_14918997 | 0.05 |
ENSDART00000164119
|
diabloa
|
diablo, IAP-binding mitochondrial protein a |
| chr16_+_17706214 | 0.05 |
|
|
|
| chr4_-_4241386 | 0.04 |
ENSDART00000103316
|
cd9b
|
CD9 molecule b |
| chr3_+_40265548 | 0.04 |
ENSDART00000103486
|
tnrc18
|
trinucleotide repeat containing 18 |
| chr13_-_9543393 | 0.04 |
ENSDART00000041609
|
tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr16_+_24047736 | 0.03 |
|
|
|
| chr18_+_41242323 | 0.03 |
ENSDART00000138552
ENSDART00000145863 |
trip12
|
thyroid hormone receptor interactor 12 |
| chr15_-_34700453 | 0.03 |
ENSDART00000140882
|
ispd
|
isoprenoid synthase domain containing |
| chr9_+_1363455 | 0.02 |
ENSDART00000140917
|
prkra
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr18_-_14932910 | 0.02 |
ENSDART00000145842
|
trabd
|
TraB domain containing |
| chr5_+_58017820 | 0.02 |
ENSDART00000083015
|
ccdc84
|
coiled-coil domain containing 84 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.3 | 2.4 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.3 | 0.9 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.3 | 0.9 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.3 | 1.1 | GO:0045777 | positive regulation of blood pressure(GO:0045777) |
| 0.2 | 1.2 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.2 | 0.5 | GO:0048714 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.2 | 1.0 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.2 | 1.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.2 | 0.8 | GO:1901976 | regulation of cell cycle checkpoint(GO:1901976) |
| 0.1 | 0.7 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.1 | 1.4 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.1 | 1.2 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.1 | 0.7 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.4 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 0.6 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 1.2 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 0.6 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 1.4 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 1.9 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.1 | 0.5 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 1.1 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 1.0 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 0.6 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 1.7 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 0.9 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.1 | 0.6 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 1.9 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.1 | 0.8 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 0.4 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.3 | GO:0050832 | neutrophil mediated immunity(GO:0002446) defense response to fungus(GO:0050832) |
| 0.1 | 0.6 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.0 | 0.4 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.3 | GO:0033183 | regulation of histone ubiquitination(GO:0033182) negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.3 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.9 | GO:2000779 | regulation of double-strand break repair(GO:2000779) |
| 0.0 | 0.4 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.3 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.3 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.5 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.7 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.3 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.0 | 0.4 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.7 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.2 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.3 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.3 | GO:0060029 | convergent extension involved in organogenesis(GO:0060029) |
| 0.0 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 0.7 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.2 | 0.8 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.5 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.1 | 0.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.8 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.6 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 0.3 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.6 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.0 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.1 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.3 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 1.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.3 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 1.4 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.6 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 1.5 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.0 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.8 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0033971 | hydroxyisourate hydrolase activity(GO:0033971) |
| 0.3 | 0.9 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 0.6 | GO:0004482 | mRNA (guanine-N7-)-methyltransferase activity(GO:0004482) |
| 0.2 | 0.7 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.2 | 1.7 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.6 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 0.6 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.6 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 2.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 1.2 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 1.0 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.1 | 0.4 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.1 | 1.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.2 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.1 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 2.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 1.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.4 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 1.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.0 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.6 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.6 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 1.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.6 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 1.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.7 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |