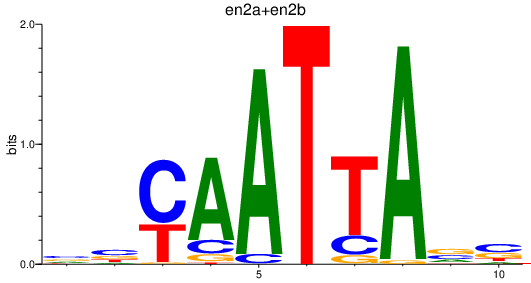
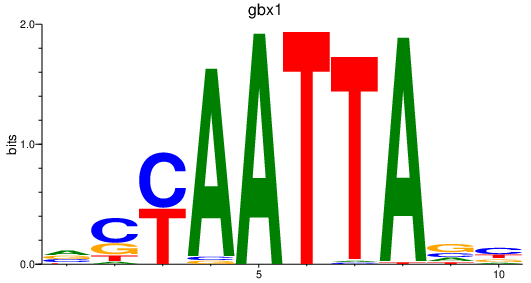
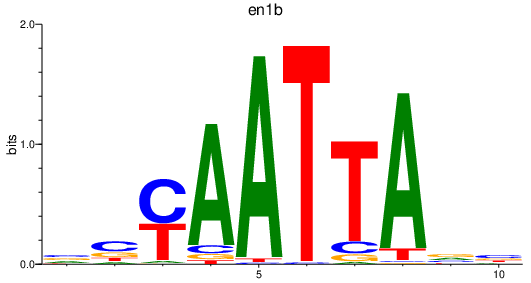
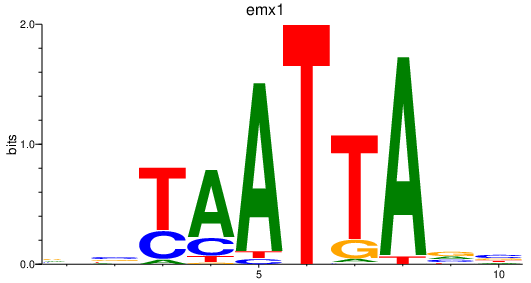
Project
DANIO-CODE
Navigation
Downloads
Results for en2a+en2b_gbx1_en1b_emx1
Z-value: 1.64
Transcription factors associated with en2a+en2b_gbx1_en1b_emx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
en2a
|
ENSDARG00000026599 | engrailed homeobox 2a |
|
en2b
|
ENSDARG00000038868 | engrailed homeobox 2b |
|
gbx1
|
ENSDARG00000071418 | gastrulation brain homeobox 1 |
|
en1b
|
ENSDARG00000098730 | engrailed homeobox 1b |
|
emx1
|
ENSDARG00000039569 | empty spiracles homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gbx1 | dr10_dc_chr24_+_34227202_34227285 | -0.91 | 1.4e-06 | Click! |
| en2b | dr10_dc_chr2_+_29992879_29992897 | -0.87 | 1.2e-05 | Click! |
| emx1 | dr10_dc_chr13_+_14873339_14873426 | -0.78 | 3.6e-04 | Click! |
| en2a | dr10_dc_chr7_-_40713381_40713426 | -0.77 | 4.6e-04 | Click! |
| en1b | dr10_dc_chr1_+_6995893_6995939 | -0.76 | 5.7e-04 | Click! |
Activity profile of en2a+en2b_gbx1_en1b_emx1 motif
Sorted Z-values of en2a+en2b_gbx1_en1b_emx1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of en2a+en2b_gbx1_en1b_emx1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_25740782 | 15.97 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr8_-_23759076 | 10.93 |
ENSDART00000145894
|
zgc:195245
|
zgc:195245 |
| chr11_-_6442588 | 9.77 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr24_-_14446593 | 9.26 |
|
|
|
| chr18_-_40718244 | 9.24 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr10_-_34971985 | 8.69 |
ENSDART00000141201
|
ccna1
|
cyclin A1 |
| chr10_-_21404605 | 7.87 |
ENSDART00000125167
|
avd
|
avidin |
| chr10_-_34058331 | 7.86 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr11_-_44539778 | 7.86 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr1_-_54570813 | 7.66 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr17_+_16038358 | 7.36 |
ENSDART00000155336
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr23_-_35691369 | 6.03 |
ENSDART00000142369
|
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr14_+_34150130 | 6.01 |
ENSDART00000132193
ENSDART00000141058 |
wnt8a
BX927327.1
|
wingless-type MMTV integration site family, member 8a ENSDARG00000105311 |
| KN150266v1_-_68652 | 5.98 |
|
|
|
| chr20_-_23527234 | 5.97 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr17_+_16038103 | 5.48 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| KN150456v1_-_19515 | 5.46 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr10_-_44713495 | 5.36 |
ENSDART00000076084
|
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr2_+_6341404 | 5.30 |
ENSDART00000076700
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr16_+_42567707 | 5.25 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr24_+_19270877 | 5.25 |
|
|
|
| KN150699v1_-_15078 | 5.20 |
ENSDART00000159861
|
ENSDARG00000098739
|
ENSDARG00000098739 |
| chr11_+_37371675 | 5.17 |
ENSDART00000140502
|
sh2d5
|
SH2 domain containing 5 |
| chr23_+_2786407 | 5.08 |
ENSDART00000066086
|
zgc:114123
|
zgc:114123 |
| chr24_+_8702288 | 5.07 |
ENSDART00000114810
|
sycp2l
|
synaptonemal complex protein 2-like |
| chr11_-_1524107 | 4.96 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr10_-_34971926 | 4.67 |
ENSDART00000141201
|
ccna1
|
cyclin A1 |
| KN149710v1_+_38638 | 4.59 |
|
|
|
| chr22_+_4035577 | 4.50 |
ENSDART00000170620
|
ctxn1
|
cortexin 1 |
| chr13_-_18564182 | 4.47 |
ENSDART00000176809
|
sfxn3
|
sideroflexin 3 |
| chr23_+_43868027 | 4.39 |
ENSDART00000112598
ENSDART00000169576 |
otud4
|
OTU deubiquitinase 4 |
| chr9_-_35824470 | 4.25 |
ENSDART00000140356
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr18_-_43890836 | 4.21 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr23_-_33692244 | 4.12 |
|
|
|
| chr24_+_1110206 | 4.03 |
ENSDART00000152063
|
BX005461.1
|
ENSDARG00000096447 |
| chr24_+_12689711 | 3.94 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr19_-_18664720 | 3.93 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr1_-_18118467 | 3.86 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr8_+_20108592 | 3.80 |
|
|
|
| chr14_-_8634381 | 3.77 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| chr12_-_14104939 | 3.67 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr21_-_32027717 | 3.67 |
ENSDART00000131651
|
ENSDARG00000073961
|
ENSDARG00000073961 |
| chr6_+_18941135 | 3.58 |
ENSDART00000044519
|
cx44.2
|
connexin 44.2 |
| chr7_+_24257251 | 3.49 |
ENSDART00000136473
|
ENSDARG00000079281
|
ENSDARG00000079281 |
| chr10_+_6925373 | 3.44 |
ENSDART00000128866
|
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr22_+_17803309 | 3.34 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr13_-_37530793 | 3.33 |
ENSDART00000141295
|
si:dkey-188i13.11
|
si:dkey-188i13.11 |
| chr12_+_46281592 | 3.20 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr2_-_38305602 | 3.15 |
ENSDART00000061677
|
si:ch211-14a17.6
|
si:ch211-14a17.6 |
| chr20_-_14218080 | 3.14 |
ENSDART00000104032
|
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr10_-_32550351 | 3.13 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr23_+_28396415 | 3.11 |
ENSDART00000142179
|
birc5b
|
baculoviral IAP repeat containing 5b |
| chr1_-_22617455 | 3.10 |
ENSDART00000137567
|
smim14
|
small integral membrane protein 14 |
| chr5_+_29193876 | 3.10 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr2_-_57020663 | 3.09 |
|
|
|
| KN150266v1_-_68620 | 2.99 |
|
|
|
| chr11_-_44756789 | 2.97 |
ENSDART00000161712
|
syngr2b
|
synaptogyrin 2b |
| chr7_+_72925910 | 2.95 |
ENSDART00000175267
ENSDART00000179638 |
CABZ01067170.1
|
ENSDARG00000106453 |
| chr5_-_14783255 | 2.93 |
ENSDART00000052712
|
gnb1l
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
| chr5_+_6391432 | 2.93 |
ENSDART00000170564
ENSDART00000086666 |
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr10_-_35313462 | 2.92 |
ENSDART00000139107
|
prr11
|
proline rich 11 |
| chr20_-_6542402 | 2.88 |
ENSDART00000054653
|
mcm3l
|
MCM3 minichromosome maintenance deficient 3 (S. cerevisiae), like |
| chr22_+_8973724 | 2.85 |
ENSDART00000106414
|
rnh1
|
ribonuclease/angiogenin inhibitor 1 |
| chr23_-_32267833 | 2.78 |
|
|
|
| chr10_-_44713414 | 2.75 |
ENSDART00000076084
|
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr24_+_39630741 | 2.72 |
ENSDART00000132939
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr11_-_6442490 | 2.71 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr8_-_1833095 | 2.67 |
ENSDART00000114476
ENSDART00000091235 ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr12_+_47448318 | 2.63 |
ENSDART00000152857
|
fmn2b
|
formin 2b |
| chr3_+_28729443 | 2.62 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr6_-_2000017 | 2.61 |
ENSDART00000158535
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr10_+_19625897 | 2.58 |
|
|
|
| KN149735v1_-_1679 | 2.57 |
|
|
|
| chr15_-_16241412 | 2.56 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr4_-_26106485 | 2.55 |
ENSDART00000100611
|
si:ch211-244b2.3
|
si:ch211-244b2.3 |
| chr10_+_35313772 | 2.49 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr24_-_19573615 | 2.47 |
ENSDART00000158952
ENSDART00000109107 ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr19_+_21209328 | 2.44 |
ENSDART00000142463
|
txnl4a
|
thioredoxin-like 4A |
| chr20_-_29961498 | 2.38 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr16_+_42567668 | 2.33 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr20_-_29961589 | 2.32 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr18_+_20571619 | 2.32 |
ENSDART00000040074
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr24_+_12689887 | 2.31 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr4_+_4825461 | 2.27 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr2_-_27892824 | 2.26 |
|
|
|
| chr12_+_46281623 | 2.26 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr13_-_37530730 | 2.22 |
ENSDART00000141295
|
si:dkey-188i13.11
|
si:dkey-188i13.11 |
| chr5_+_36168475 | 2.21 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr7_-_51497945 | 2.18 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr10_-_25448712 | 2.15 |
ENSDART00000140023
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr20_+_14218237 | 2.14 |
ENSDART00000044937
|
kcns3b
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3b |
| chr6_-_3821922 | 2.14 |
ENSDART00000171944
|
slc25a12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr3_+_42383724 | 2.14 |
|
|
|
| chr11_+_30005768 | 2.11 |
ENSDART00000167618
|
CR790368.1
|
ENSDARG00000100936 |
| chr9_+_33310981 | 2.09 |
|
|
|
| chr3_-_29725539 | 2.09 |
|
|
|
| chr17_+_7377230 | 2.08 |
ENSDART00000157123
|
shprh
|
SNF2 histone linker PHD RING helicase |
| chr12_+_46281511 | 2.05 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr7_+_20092235 | 2.03 |
ENSDART00000139274
|
ponzr1
|
plac8 onzin related protein 1 |
| chr19_+_31998646 | 2.02 |
ENSDART00000132182
|
gmnn
|
geminin, DNA replication inhibitor |
| chr12_+_22552867 | 2.00 |
ENSDART00000152930
|
cdca9
|
cell division cycle associated 9 |
| chr17_-_25630635 | 1.99 |
ENSDART00000149060
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr16_-_42105636 | 1.98 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr20_-_20425034 | 1.98 |
ENSDART00000152795
|
snapc1a
|
small nuclear RNA activating complex, polypeptide 1a |
| chr8_-_44247277 | 1.94 |
|
|
|
| chr17_-_45021393 | 1.93 |
|
|
|
| chr16_-_41764849 | 1.92 |
ENSDART00000084610
|
cep85
|
centrosomal protein 85 |
| chr1_+_35253862 | 1.92 |
ENSDART00000139636
|
zgc:152968
|
zgc:152968 |
| chr3_-_15060685 | 1.91 |
ENSDART00000037906
|
hirip3
|
HIRA interacting protein 3 |
| chr17_+_16421892 | 1.90 |
ENSDART00000136078
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr5_+_37303599 | 1.88 |
ENSDART00000097754
ENSDART00000162470 |
tmprss4b
|
transmembrane protease, serine 4b |
| chr21_-_39521698 | 1.88 |
ENSDART00000020174
|
dynll2b
|
dynein, light chain, LC8-type 2b |
| chr19_-_5186692 | 1.86 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr14_-_9686650 | 1.86 |
|
|
|
| chr7_+_20091996 | 1.86 |
ENSDART00000138786
|
ponzr1
|
plac8 onzin related protein 1 |
| chr2_-_15656155 | 1.85 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr6_+_28218420 | 1.84 |
ENSDART00000171216
ENSDART00000171377 |
si:ch73-14h10.2
|
si:ch73-14h10.2 |
| chr14_+_23420053 | 1.84 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr8_+_11287550 | 1.83 |
ENSDART00000115057
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr24_-_19573966 | 1.82 |
ENSDART00000158952
ENSDART00000109107 ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr2_+_6341345 | 1.79 |
ENSDART00000058256
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr9_+_18821640 | 1.79 |
ENSDART00000006514
|
gtf2f2b
|
general transcription factor IIF, polypeptide 2b |
| chr10_+_16078021 | 1.74 |
ENSDART00000065037
ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr11_-_12024136 | 1.74 |
ENSDART00000111919
|
sp2
|
sp2 transcription factor |
| chr5_+_32687626 | 1.74 |
ENSDART00000146759
|
med22
|
mediator complex subunit 22 |
| chr24_+_16402587 | 1.73 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr19_+_38262119 | 1.72 |
ENSDART00000042276
|
nxph1
|
neurexophilin 1 |
| chr24_-_9838947 | 1.72 |
ENSDART00000138576
|
zgc:171977
|
zgc:171977 |
| chr12_-_33256671 | 1.71 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr17_+_10582214 | 1.70 |
ENSDART00000051527
|
tbpl2
|
TATA box binding protein like 2 |
| chr8_+_49128711 | 1.69 |
ENSDART00000079631
|
rad21l1
|
RAD21 cohesin complex component like 1 |
| chr25_-_13394261 | 1.68 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr15_+_28477741 | 1.68 |
ENSDART00000057257
|
pitpnaa
|
phosphatidylinositol transfer protein, alpha a |
| chr8_-_32376710 | 1.66 |
ENSDART00000098850
|
lipg
|
lipase, endothelial |
| chr6_+_40925259 | 1.65 |
ENSDART00000002728
ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr11_-_44539726 | 1.65 |
ENSDART00000173360
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr2_+_37194276 | 1.63 |
ENSDART00000048277
|
copa
|
coatomer protein complex, subunit alpha |
| chr7_+_37818365 | 1.63 |
ENSDART00000052365
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr12_+_3632548 | 1.61 |
|
|
|
| chr10_-_2944190 | 1.60 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr25_+_33486711 | 1.59 |
ENSDART00000121498
|
ice2
|
interactor of little elongator complex ELL subunit 2 |
| chr12_+_22459177 | 1.57 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr12_-_33256754 | 1.56 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr10_-_25808063 | 1.53 |
ENSDART00000134176
|
postna
|
periostin, osteoblast specific factor a |
| chr24_-_14446522 | 1.50 |
|
|
|
| chr1_+_518777 | 1.49 |
ENSDART00000109083
|
txnl4b
|
thioredoxin-like 4B |
| chr19_+_40558066 | 1.49 |
ENSDART00000049968
|
si:ch211-173p18.3
|
si:ch211-173p18.3 |
| chr24_-_34794538 | 1.49 |
ENSDART00000171009
ENSDART00000170046 |
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr14_+_23419864 | 1.47 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr19_+_42657913 | 1.47 |
ENSDART00000131574
ENSDART00000135436 |
jtb
|
jumping translocation breakpoint |
| chr8_-_21039978 | 1.45 |
ENSDART00000137606
ENSDART00000146532 |
zgc:112962
|
zgc:112962 |
| chr6_-_37766424 | 1.45 |
ENSDART00000149722
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr14_-_33605295 | 1.43 |
ENSDART00000168546
|
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr17_+_10582044 | 1.43 |
ENSDART00000051527
|
tbpl2
|
TATA box binding protein like 2 |
| chr18_-_49291686 | 1.42 |
ENSDART00000174038
|
si:zfos-464b6.2
|
si:zfos-464b6.2 |
| chr14_+_43357313 | 1.42 |
|
|
|
| chr24_-_24999240 | 1.41 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr8_-_21110262 | 1.41 |
ENSDART00000143192
|
cpt2
|
carnitine palmitoyltransferase 2 |
| chr19_-_8849482 | 1.40 |
ENSDART00000170416
|
si:ch73-350k19.1
|
si:ch73-350k19.1 |
| chr13_-_10214396 | 1.40 |
ENSDART00000132231
|
AL929457.1
|
ENSDARG00000095483 |
| chr2_+_10209233 | 1.39 |
ENSDART00000160304
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr14_-_14353451 | 1.39 |
ENSDART00000170355
ENSDART00000159888 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr1_+_18118735 | 1.38 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr19_-_33264349 | 1.37 |
|
|
|
| chr3_-_26052785 | 1.37 |
ENSDART00000147517
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr3_+_38497816 | 1.37 |
|
|
|
| chr16_+_47283253 | 1.37 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr18_+_20571460 | 1.36 |
ENSDART00000151990
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr20_+_27188382 | 1.36 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr19_-_20819477 | 1.36 |
ENSDART00000151356
|
dazl
|
deleted in azoospermia-like |
| chr22_+_25734113 | 1.35 |
ENSDART00000136334
|
si:ch211-250e5.16
|
si:ch211-250e5.16 |
| chr3_-_15060501 | 1.33 |
ENSDART00000037906
|
hirip3
|
HIRA interacting protein 3 |
| chr15_+_6462447 | 1.33 |
ENSDART00000065824
|
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr15_-_25164321 | 1.32 |
ENSDART00000154628
|
fam101b
|
family with sequence similarity 101, member B |
| chr20_+_37023072 | 1.32 |
ENSDART00000155058
|
CR388421.1
|
ENSDARG00000096706 |
| chr12_+_33257120 | 1.31 |
|
|
|
| chr25_-_9889107 | 1.29 |
ENSDART00000137407
|
AL929493.1
|
ENSDARG00000093575 |
| chr5_+_19429620 | 1.29 |
ENSDART00000088819
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr16_-_24727689 | 1.29 |
ENSDART00000167121
|
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr2_+_55065658 | 1.29 |
|
|
|
| chr24_-_36721857 | 1.28 |
|
|
|
| chr11_+_23799984 | 1.28 |
|
|
|
| chr2_+_42923652 | 1.28 |
ENSDART00000168318
|
BX005083.1
|
ENSDARG00000097562 |
| chr24_-_14447519 | 1.25 |
|
|
|
| chr12_-_26943743 | 1.25 |
|
|
|
| chr11_+_12754166 | 1.24 |
ENSDART00000123445
ENSDART00000163364 ENSDART00000066122 |
rtel1
|
regulator of telomere elongation helicase 1 |
| chr12_-_33257026 | 1.24 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr9_-_32348003 | 1.24 |
ENSDART00000037182
|
ankrd44
|
ankyrin repeat domain 44 |
| chr9_-_32347822 | 1.24 |
ENSDART00000037182
|
ankrd44
|
ankyrin repeat domain 44 |
| chr8_+_45326435 | 1.24 |
ENSDART00000134161
|
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr1_-_17000212 | 1.22 |
ENSDART00000146258
|
cfap97
|
cilia and flagella associated protein 97 |
| chr19_+_15536640 | 1.21 |
ENSDART00000098970
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr20_-_14218236 | 1.21 |
ENSDART00000168434
|
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr2_+_3115593 | 1.20 |
ENSDART00000160715
|
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr10_+_516924 | 1.19 |
ENSDART00000128275
|
npffr1l3
|
neuropeptide FF receptor 1 like 3 |
| chr23_-_2957262 | 1.19 |
ENSDART00000165955
|
zhx3
|
zinc fingers and homeoboxes 3 |
| chr14_+_8634323 | 1.18 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr19_+_1743359 | 1.17 |
ENSDART00000166744
|
dennd3a
|
DENN/MADD domain containing 3a |
| chr8_-_25015215 | 1.16 |
ENSDART00000170511
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr3_-_26052601 | 1.15 |
ENSDART00000147517
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.5 | GO:0050957 | equilibrioception(GO:0050957) |
| 1.8 | 7.2 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 1.7 | 6.8 | GO:0072111 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 1.3 | 5.1 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 1.3 | 6.3 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 1.2 | 4.9 | GO:0070986 | left/right axis specification(GO:0070986) |
| 1.1 | 4.4 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 1.0 | 3.1 | GO:0051311 | meiotic metaphase plate congression(GO:0051311) |
| 0.9 | 3.6 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.8 | 2.5 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.8 | 13.3 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.6 | 1.9 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.6 | 15.4 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.6 | 5.6 | GO:0061458 | gonad development(GO:0008406) reproductive structure development(GO:0048608) reproductive system development(GO:0061458) |
| 0.5 | 3.9 | GO:0072088 | renal tubule morphogenesis(GO:0061333) nephron tubule morphogenesis(GO:0072078) nephron epithelium morphogenesis(GO:0072088) |
| 0.4 | 1.2 | GO:0090656 | t-circle formation(GO:0090656) |
| 0.4 | 2.9 | GO:1902315 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.4 | 1.9 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.4 | 1.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.3 | 5.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.3 | 6.8 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.3 | 1.8 | GO:0035264 | multicellular organism growth(GO:0035264) |
| 0.3 | 2.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.3 | 1.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.3 | 7.1 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.3 | 4.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 1.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.3 | 1.9 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.3 | 1.4 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.3 | 1.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) mRNA pseudouridine synthesis(GO:1990481) |
| 0.3 | 14.8 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.2 | 2.1 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.2 | 0.9 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.2 | 0.5 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.2 | 0.7 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.2 | 1.7 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.2 | 0.6 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.2 | 3.6 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.2 | 0.6 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.2 | 0.9 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.2 | 5.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.2 | 1.9 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.2 | 0.9 | GO:1900182 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.2 | 1.0 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.2 | 3.0 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.2 | 3.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.2 | 0.5 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.2 | 1.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.2 | 1.9 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.2 | 0.9 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 1.0 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 0.7 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.4 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.1 | 0.9 | GO:1900186 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.4 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.1 | 2.0 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 1.3 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) melanocyte migration(GO:0097324) |
| 0.1 | 1.8 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.6 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.4 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 5.4 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 2.7 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 1.6 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.1 | 0.8 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.1 | 3.1 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.1 | 1.6 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 1.6 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 0.3 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 1.0 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 0.2 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.1 | 2.5 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 3.3 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 0.3 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 5.5 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.1 | 0.6 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.1 | 0.4 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) |
| 0.1 | 0.4 | GO:0043090 | amino acid import(GO:0043090) L-amino acid import(GO:0043092) |
| 0.1 | 0.6 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 0.7 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.1 | 3.0 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.5 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) skin epidermis development(GO:0098773) |
| 0.1 | 1.0 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.9 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 0.9 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.4 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) renal absorption(GO:0070293) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 4.5 | GO:1990542 | mitochondrial transmembrane transport(GO:1990542) |
| 0.1 | 0.5 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.4 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.3 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.3 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 5.7 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.1 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.3 | GO:0046098 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine metabolic process(GO:0046098) guanine biosynthetic process(GO:0046099) hypoxanthine metabolic process(GO:0046100) |
| 0.1 | 0.4 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 6.0 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.1 | 0.6 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 0.4 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 0.8 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 2.6 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.1 | 1.7 | GO:0072329 | monocarboxylic acid catabolic process(GO:0072329) |
| 0.1 | 0.4 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 0.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.7 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 1.1 | GO:0030301 | cholesterol transport(GO:0030301) |
| 0.0 | 0.7 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.2 | GO:0019755 | one-carbon compound transport(GO:0019755) |
| 0.0 | 0.2 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.7 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 1.2 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.6 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 4.8 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.9 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 1.8 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.4 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.2 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 1.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.8 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 2.5 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 1.1 | GO:0071230 | response to amino acid(GO:0043200) cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 2.6 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.0 | 1.7 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.0 | 0.2 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 1.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 2.1 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 1.2 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.4 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.9 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.3 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.0 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.5 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 1.0 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.2 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0030817 | regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 1.5 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.2 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.0 | 0.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 2.3 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.8 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 1.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 1.2 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.5 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.4 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.4 | GO:1903321 | negative regulation of protein ubiquitination(GO:0031397) negative regulation of protein modification by small protein conjugation or removal(GO:1903321) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.5 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 0.6 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.5 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.6 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.4 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.7 | GO:0021536 | diencephalon development(GO:0021536) |
| 0.0 | 0.4 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 2.6 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.3 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.6 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.2 | GO:0046460 | triglyceride biosynthetic process(GO:0019432) neutral lipid biosynthetic process(GO:0046460) acylglycerol biosynthetic process(GO:0046463) |
| 0.0 | 0.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.1 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 1.0 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 1.2 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 0.3 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.3 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.3 | GO:0030217 | T cell differentiation(GO:0030217) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 13.4 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 1.7 | 6.8 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.7 | 2.0 | GO:0019185 | snRNA-activating protein complex(GO:0019185) |
| 0.6 | 1.7 | GO:0034990 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.5 | 11.1 | GO:0043186 | P granule(GO:0043186) |
| 0.5 | 1.4 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.4 | 1.8 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.4 | 2.6 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.3 | 4.8 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.3 | 0.9 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.2 | 4.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 16.6 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.2 | 2.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 2.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 2.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 1.0 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.2 | 0.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.2 | 3.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 1.0 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 0.9 | GO:0000811 | GINS complex(GO:0000811) |
| 0.2 | 1.4 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.2 | 2.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 4.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 1.1 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 1.6 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.1 | 0.9 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 4.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 2.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 0.9 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 3.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 2.0 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 0.7 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 8.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 4.3 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 3.5 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 1.2 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.1 | 0.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 3.2 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 2.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 1.5 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 1.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 1.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 8.3 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.6 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 9.5 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.6 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.6 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 4.4 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 1.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.6 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.7 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 1.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 5.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 12.9 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 1.6 | 7.9 | GO:0009374 | biotin binding(GO:0009374) |
| 1.1 | 2.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.9 | 6.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.8 | 7.1 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.8 | 6.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.6 | 4.5 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.6 | 1.9 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.6 | 1.7 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.5 | 1.6 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.5 | 2.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.5 | 5.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.4 | 2.7 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.4 | 1.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.4 | 3.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.4 | 2.6 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.4 | 2.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.3 | 9.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.3 | 1.7 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.3 | 1.8 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.2 | 3.1 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.2 | 1.9 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.2 | 15.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 0.7 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 1.3 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.2 | 0.6 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 3.6 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.2 | 2.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 2.2 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.2 | 1.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 0.7 | GO:1902387 | ceramide transporter activity(GO:0035620) sphingolipid transporter activity(GO:0046624) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.2 | 1.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 2.1 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.2 | 0.9 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 1.0 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.2 | 3.1 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.2 | 1.0 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 1.6 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.2 | 0.5 | GO:0097001 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.1 | 0.4 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 5.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.7 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 4.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.9 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 6.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 1.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.4 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.1 | 0.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.4 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 0.9 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.2 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 24.0 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.1 | 0.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 2.9 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.8 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.7 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 5.6 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 2.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 0.9 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 0.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 6.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.3 | GO:0052657 | guanine phosphoribosyltransferase activity(GO:0052657) |
| 0.1 | 0.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 2.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 6.5 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 1.0 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 0.4 | GO:0019869 | chloride channel regulator activity(GO:0017081) chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.5 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 1.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.5 | GO:0015368 | calcium:cation antiporter activity(GO:0015368) |
| 0.1 | 0.7 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.3 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 2.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 1.1 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 2.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 4.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 5.5 | GO:0015103 | inorganic anion transmembrane transporter activity(GO:0015103) |
| 0.1 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 1.8 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 0.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 1.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.0 | 0.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.4 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 4.8 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 6.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 2.1 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 1.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.4 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.4 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.8 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 4.6 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 2.4 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 1.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.1 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 1.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 3.5 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 1.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.0 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.0 | 1.6 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.4 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.1 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.6 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 13.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 3.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 2.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 0.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 0.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 2.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 1.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 0.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 7.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 1.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 2.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 1.6 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 1.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.7 | 13.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.3 | 1.0 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 2.6 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.2 | 0.9 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 1.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 3.0 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.2 | 2.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 0.1 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.1 | 1.2 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 1.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 2.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 3.1 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 1.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 0.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 3.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 2.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.3 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.6 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.7 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | Genes involved in TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |