Project
DANIO-CODE
Navigation
Downloads
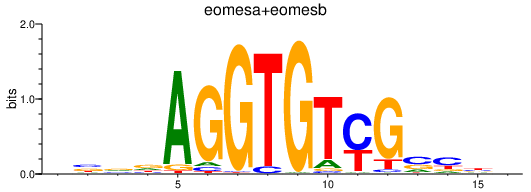
Results for eomesa+eomesb
Z-value: 0.52
Transcription factors associated with eomesa+eomesb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
eomesa
|
ENSDARG00000006640 | eomesodermin homolog a |
|
eomesb
|
ENSDARG00000104243 | eomesodermin homolog b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| eomesa | dr10_dc_chr19_-_867262_867279 | -0.45 | 8.1e-02 | Click! |
Activity profile of eomesa+eomesb motif
Sorted Z-values of eomesa+eomesb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of eomesa+eomesb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_44006939 | 1.70 |
|
|
|
| chr22_-_36721626 | 0.73 |
ENSDART00000017188
ENSDART00000124698 |
ncl
|
nucleolin |
| chr22_+_25113293 | 0.72 |
ENSDART00000171851
|
si:ch211-226h8.4
|
si:ch211-226h8.4 |
| chr7_+_23782982 | 0.64 |
|
|
|
| chr1_-_55072271 | 0.61 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr11_+_18823629 | 0.60 |
|
|
|
| chr13_+_7607232 | 0.55 |
|
|
|
| KN149861v1_-_5655 | 0.54 |
|
|
|
| chr16_+_4825665 | 0.52 |
|
|
|
| chr11_-_1524107 | 0.48 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr22_-_18519706 | 0.44 |
ENSDART00000132156
|
cirbpb
|
cold inducible RNA binding protein b |
| chr22_+_25156826 | 0.39 |
ENSDART00000140670
ENSDART00000105308 |
si:ch211-226h8.4
|
si:ch211-226h8.4 |
| chr10_-_25246786 | 0.38 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr22_+_25734113 | 0.37 |
ENSDART00000136334
|
si:ch211-250e5.16
|
si:ch211-250e5.16 |
| chr5_-_15971338 | 0.37 |
ENSDART00000110437
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr17_+_23534953 | 0.37 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr16_+_25192196 | 0.35 |
ENSDART00000033211
|
zgc:66448
|
zgc:66448 |
| chr2_+_2977250 | 0.35 |
|
|
|
| chr8_-_43151763 | 0.35 |
ENSDART00000134801
|
ccdc92
|
coiled-coil domain containing 92 |
| chr1_-_39297624 | 0.35 |
ENSDART00000165373
|
cdk2ap2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr16_+_24046996 | 0.34 |
|
|
|
| chr11_+_18823762 | 0.33 |
|
|
|
| chr6_-_34951317 | 0.32 |
|
|
|
| chr1_-_39297686 | 0.31 |
ENSDART00000165373
|
cdk2ap2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr5_+_61656465 | 0.31 |
ENSDART00000130177
ENSDART00000050879 |
pho
|
phoenix |
| chr5_-_41044841 | 0.31 |
ENSDART00000051092
|
riok2
|
RIO kinase 2 (yeast) |
| chr17_-_27030048 | 0.29 |
ENSDART00000050018
|
cnksr1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr17_-_33463210 | 0.27 |
ENSDART00000135218
|
ccdc28a
|
coiled-coil domain containing 28A |
| chr22_+_25754857 | 0.27 |
ENSDART00000174421
|
AL929192.1
|
ENSDARG00000106684 |
| chr8_+_25988813 | 0.26 |
ENSDART00000058100
|
xpc
|
xeroderma pigmentosum, complementation group C |
| chr18_+_3435932 | 0.26 |
|
|
|
| chr6_-_34951155 | 0.23 |
|
|
|
| chr22_+_25755073 | 0.23 |
ENSDART00000174421
|
AL929192.1
|
ENSDARG00000106684 |
| chr11_+_18856937 | 0.23 |
|
|
|
| chr19_+_40439358 | 0.21 |
ENSDART00000123647
|
sfpq
|
splicing factor proline/glutamine-rich |
| chr10_+_6339177 | 0.21 |
ENSDART00000160813
|
tpm2
|
tropomyosin 2 (beta) |
| chr2_+_2976671 | 0.21 |
ENSDART00000081976
|
rock1
|
Rho-associated, coiled-coil containing protein kinase 1 |
| chr5_+_30436723 | 0.21 |
ENSDART00000141463
|
zzef1
|
zinc finger, ZZ-type with EF hand domain 1 |
| chr2_+_22936475 | 0.20 |
ENSDART00000126891
ENSDART00000130603 |
sept2
|
septin 2 |
| chr10_+_42841562 | 0.20 |
|
|
|
| chr1_-_55072375 | 0.20 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr22_+_25754948 | 0.18 |
ENSDART00000174421
|
AL929192.1
|
ENSDARG00000106684 |
| chr12_+_35855929 | 0.16 |
|
|
|
| chr16_+_4825019 | 0.16 |
ENSDART00000167665
|
LIN28A
|
lin-28 homolog A |
| chr10_+_6339059 | 0.16 |
ENSDART00000160813
|
tpm2
|
tropomyosin 2 (beta) |
| chr4_-_11604184 | 0.16 |
ENSDART00000170136
|
net1
|
neuroepithelial cell transforming 1 |
| chr12_-_17302222 | 0.15 |
ENSDART00000079144
|
ptenb
|
phosphatase and tensin homolog B |
| chr12_-_622463 | 0.15 |
ENSDART00000158563
|
wu:fj29h11
|
wu:fj29h11 |
| chr22_-_4867538 | 0.14 |
ENSDART00000081801
|
ncl1
|
nicalin |
| KN149861v1_-_5543 | 0.14 |
|
|
|
| chr11_-_40239946 | 0.14 |
ENSDART00000165394
|
si:dkeyp-61b2.1
|
si:dkeyp-61b2.1 |
| chr7_+_30222432 | 0.14 |
ENSDART00000027466
|
fam63b
|
family with sequence similarity 63, member B |
| chr15_-_45522055 | 0.14 |
ENSDART00000113494
|
MB21D2
|
Mab-21 domain containing 2 |
| chr10_+_6338988 | 0.14 |
ENSDART00000160813
|
tpm2
|
tropomyosin 2 (beta) |
| chr22_-_18520332 | 0.14 |
|
|
|
| chr18_+_50921419 | 0.14 |
ENSDART00000058468
ENSDART00000008696 |
cttn
|
cortactin |
| KN150593v1_+_10332 | 0.13 |
|
|
|
| chr3_-_44113070 | 0.12 |
ENSDART00000160717
|
znf750
|
zinc finger protein 750 |
| chr7_-_14835871 | 0.12 |
ENSDART00000172489
|
CU280645.1
|
ENSDARG00000098326 |
| chr5_-_41045017 | 0.12 |
ENSDART00000051092
|
riok2
|
RIO kinase 2 (yeast) |
| chr6_-_34951506 | 0.12 |
|
|
|
| chr17_-_33463482 | 0.11 |
ENSDART00000164064
|
ccdc28a
|
coiled-coil domain containing 28A |
| chr9_-_24013313 | 0.11 |
ENSDART00000059209
|
wdfy2
|
WD repeat and FYVE domain containing 2 |
| chr8_-_43151611 | 0.11 |
ENSDART00000134801
|
ccdc92
|
coiled-coil domain containing 92 |
| chr9_-_12913920 | 0.11 |
ENSDART00000122799
|
ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr11_-_40464213 | 0.11 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr3_+_36941624 | 0.11 |
ENSDART00000125611
|
retreg3
|
reticulophagy regulator family member 3 |
| chr22_-_18520027 | 0.11 |
ENSDART00000132156
|
cirbpb
|
cold inducible RNA binding protein b |
| chr25_-_23485520 | 0.11 |
ENSDART00000149107
|
nap1l4a
|
nucleosome assembly protein 1-like 4a |
| chr17_+_23535117 | 0.08 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr2_+_2976973 | 0.08 |
ENSDART00000163587
|
rock1
|
Rho-associated, coiled-coil containing protein kinase 1 |
| chr3_+_62116865 | 0.08 |
ENSDART00000112428
|
iqck
|
IQ motif containing K |
| chr20_-_7594437 | 0.06 |
|
|
|
| chr5_-_41044810 | 0.05 |
ENSDART00000051092
|
riok2
|
RIO kinase 2 (yeast) |
| chr21_-_14719281 | 0.05 |
ENSDART00000067004
|
phpt1
|
phosphohistidine phosphatase 1 |
| chr24_-_55232 | 0.04 |
|
|
|
| chr17_-_29295450 | 0.04 |
ENSDART00000133668
|
tecpr2
|
tectonin beta-propeller repeat containing 2 |
| chr24_-_36307897 | 0.03 |
ENSDART00000154395
|
esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr11_-_40464253 | 0.02 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr8_-_47855945 | 0.02 |
ENSDART00000145429
|
si:dkeyp-104h9.5
|
si:dkeyp-104h9.5 |
| chr11_+_14254556 | 0.02 |
ENSDART00000102518
|
si:ch211-262i1.5
|
si:ch211-262i1.5 |
| chr24_-_36307855 | 0.01 |
ENSDART00000111891
|
esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr8_-_19167286 | 0.00 |
ENSDART00000161646
|
sema6bb
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Bb |
| chr1_-_55072064 | 0.00 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.1 | 0.3 | GO:0070655 | mechanosensory epithelium regeneration(GO:0070655) mechanoreceptor differentiation involved in mechanosensory epithelium regeneration(GO:0070656) neuromast regeneration(GO:0070657) neuromast hair cell differentiation involved in neuromast regeneration(GO:0070658) |
| 0.1 | 0.3 | GO:1903392 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 | 0.7 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.2 | GO:0021547 | midbrain-hindbrain boundary initiation(GO:0021547) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.8 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.2 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.5 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.2 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.5 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.3 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |