Project
DANIO-CODE
Navigation
Downloads
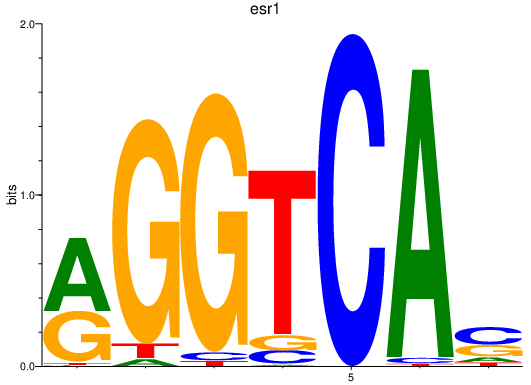
Results for esr1
Z-value: 1.16
Transcription factors associated with esr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
esr1
|
ENSDARG00000004111 | estrogen receptor 1 |
Activity profile of esr1 motif
Sorted Z-values of esr1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of esr1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_44180588 | 4.71 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr18_+_62936 | 3.59 |
ENSDART00000052638
|
slc27a2a
|
solute carrier family 27 (fatty acid transporter), member 2a |
| chr5_-_70948223 | 3.29 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr7_+_19765237 | 3.19 |
ENSDART00000100808
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr23_+_44817648 | 2.81 |
ENSDART00000143688
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr18_-_39491932 | 2.80 |
ENSDART00000122930
|
scg3
|
secretogranin III |
| chr18_-_15405161 | 2.75 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr7_+_40603904 | 2.74 |
ENSDART00000149395
|
shha
|
sonic hedgehog a |
| chr3_+_39425125 | 2.64 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr6_+_55022668 | 2.61 |
ENSDART00000158845
|
mybphb
|
myosin binding protein Hb |
| chr11_+_11217547 | 2.59 |
ENSDART00000087105
|
myom2a
|
myomesin 2a |
| chr7_+_40604000 | 2.55 |
ENSDART00000149395
|
shha
|
sonic hedgehog a |
| chr16_+_29716279 | 2.42 |
ENSDART00000137153
|
tmod4
|
tropomodulin 4 (muscle) |
| chr4_-_1346961 | 2.23 |
ENSDART00000164623
|
ptn
|
pleiotrophin |
| chr22_+_33177432 | 2.22 |
ENSDART00000126885
|
dag1
|
dystroglycan 1 |
| chr5_-_36237656 | 2.18 |
ENSDART00000032481
|
ckma
|
creatine kinase, muscle a |
| chr9_-_22528568 | 2.17 |
ENSDART00000134805
|
crygm2d1
|
crystallin, gamma M2d1 |
| chr2_-_24275851 | 2.13 |
ENSDART00000121885
|
tgfbr1a
|
transforming growth factor, beta receptor 1 a |
| chr3_+_26013873 | 2.12 |
ENSDART00000043932
|
atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr15_+_24708696 | 2.07 |
ENSDART00000043292
|
smtnl
|
smoothelin, like |
| chr9_-_53220424 | 2.03 |
|
|
|
| chr1_+_53286155 | 2.03 |
ENSDART00000139625
|
acta1a
|
actin, alpha 1a, skeletal muscle |
| chr2_-_24357532 | 1.96 |
ENSDART00000137065
|
col15a1b
|
collagen, type XV, alpha 1b |
| chr6_-_13654186 | 1.95 |
ENSDART00000150102
ENSDART00000041269 |
cryba2a
|
crystallin, beta A2a |
| chr5_-_46382728 | 1.93 |
|
|
|
| chr25_-_30818836 | 1.92 |
ENSDART00000156828
|
prr33
|
proline rich 33 |
| chr18_+_6551983 | 1.86 |
ENSDART00000160382
ENSDART00000171495 ENSDART00000160228 |
fam168a
|
family with sequence similarity 168, member A |
| chr23_-_26150495 | 1.85 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr24_-_12794057 | 1.84 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr24_-_39722595 | 1.81 |
ENSDART00000066506
|
cox6b1
|
cytochrome c oxidase subunit VIb polypeptide 1 |
| chr11_-_6051096 | 1.77 |
ENSDART00000147761
|
vsg1
|
vessel-specific 1 |
| chr12_-_43939028 | 1.76 |
ENSDART00000170723
|
zgc:112964
|
zgc:112964 |
| chr12_+_27032862 | 1.75 |
|
|
|
| chr18_+_31038010 | 1.74 |
ENSDART00000163471
|
uraha
|
urate (5-hydroxyiso-) hydrolase a |
| chr18_-_15404998 | 1.72 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr22_-_15567180 | 1.68 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr24_-_12794564 | 1.68 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr5_-_54773058 | 1.68 |
ENSDART00000145791
|
prune2
|
prune homolog 2 (Drosophila) |
| chr11_+_7148830 | 1.67 |
ENSDART00000035560
|
tmem38a
|
transmembrane protein 38A |
| chr2_+_42342148 | 1.67 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr8_-_23102555 | 1.65 |
|
|
|
| chr21_+_40082890 | 1.64 |
ENSDART00000100166
|
serpinf1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr20_-_24223659 | 1.64 |
ENSDART00000153464
|
bach2b
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2b |
| chr22_-_120677 | 1.63 |
|
|
|
| chr4_+_4841191 | 1.61 |
ENSDART00000130818
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr3_+_23587156 | 1.61 |
|
|
|
| chr16_+_24057260 | 1.60 |
ENSDART00000132742
ENSDART00000145330 |
apoc1
|
apolipoprotein C-I |
| chr14_-_136402 | 1.59 |
|
|
|
| chr22_+_11726312 | 1.59 |
ENSDART00000155366
|
krt96
|
keratin 96 |
| chr6_+_41466199 | 1.59 |
ENSDART00000177439
|
twf2a
|
twinfilin actin-binding protein 2a |
| chr2_-_28446615 | 1.56 |
ENSDART00000179495
|
cdh6
|
cadherin 6 |
| chr22_+_18218979 | 1.56 |
|
|
|
| chr10_+_4962403 | 1.56 |
ENSDART00000134679
|
CU074419.2
|
ENSDARG00000093688 |
| chr4_+_18974767 | 1.55 |
ENSDART00000066973
|
impdh1b
|
IMP (inosine 5'-monophosphate) dehydrogenase 1b |
| chr15_-_20297270 | 1.55 |
ENSDART00000123910
|
ppp1r14ab
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Ab |
| chr12_-_4648262 | 1.55 |
ENSDART00000152771
|
si:ch211-255p10.3
|
si:ch211-255p10.3 |
| chr9_-_7560915 | 1.55 |
ENSDART00000081553
|
desma
|
desmin a |
| chr23_+_6298911 | 1.54 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr10_-_22881453 | 1.54 |
|
|
|
| chr15_-_25582891 | 1.54 |
|
|
|
| chr1_+_16983775 | 1.53 |
ENSDART00000030665
|
slc25a4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr15_+_32853646 | 1.51 |
ENSDART00000167515
|
postnb
|
periostin, osteoblast specific factor b |
| chr3_+_1413640 | 1.51 |
ENSDART00000055430
|
ndufa6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 |
| chr13_-_37001997 | 1.50 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr10_-_38206238 | 1.48 |
ENSDART00000174882
|
FP016248.1
|
ENSDARG00000107804 |
| chr11_+_27970922 | 1.47 |
ENSDART00000169360
ENSDART00000043756 |
ephb2b
|
eph receptor B2b |
| chr13_-_31304614 | 1.47 |
|
|
|
| chr1_-_20218263 | 1.46 |
ENSDART00000078271
|
cpe
|
carboxypeptidase E |
| chr23_-_15574072 | 1.45 |
ENSDART00000035865
ENSDART00000104904 |
sulf2b
|
sulfatase 2b |
| chr9_-_22461196 | 1.44 |
ENSDART00000135190
|
crygm2d7
|
crystallin, gamma M2d7 |
| chr22_+_10728099 | 1.44 |
ENSDART00000145551
|
tmprss9
|
transmembrane protease, serine 9 |
| chr6_-_40724581 | 1.42 |
ENSDART00000035101
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr5_+_26840147 | 1.41 |
ENSDART00000064701
|
loxl2b
|
lysyl oxidase-like 2b |
| chr24_-_21778717 | 1.41 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr20_-_45819128 | 1.41 |
ENSDART00000124283
|
gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr10_+_6316944 | 1.41 |
ENSDART00000162428
|
tpm2
|
tropomyosin 2 (beta) |
| chr2_+_30932612 | 1.41 |
ENSDART00000132450
ENSDART00000137012 |
myom1a
|
myomesin 1a (skelemin) |
| chr11_+_30482530 | 1.40 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr3_+_47735048 | 1.40 |
ENSDART00000083024
|
gpr146
|
G protein-coupled receptor 146 |
| chr14_-_14337584 | 1.38 |
ENSDART00000167119
ENSDART00000168027 ENSDART00000167521 |
znf185
|
zinc finger protein 185 with LIM domain |
| KN150642v1_+_9419 | 1.37 |
ENSDART00000159069
|
atoh1b
|
atonal bHLH transcription factor 1b |
| chr7_-_3900223 | 1.37 |
ENSDART00000019949
|
ndrg2
|
NDRG family member 2 |
| chr11_-_29891067 | 1.36 |
ENSDART00000172106
|
scml2
|
sex comb on midleg-like 2 (Drosophila) |
| chr17_+_8874210 | 1.36 |
|
|
|
| chr21_-_19789339 | 1.36 |
|
|
|
| chr24_+_5205878 | 1.35 |
ENSDART00000106488
ENSDART00000005901 |
plod2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr20_-_17006936 | 1.34 |
ENSDART00000012859
|
psma6b
|
proteasome subunit alpha 6b |
| chr16_-_55320569 | 1.32 |
ENSDART00000156368
|
ENSDARG00000069583
|
ENSDARG00000069583 |
| chr3_-_45779817 | 1.32 |
ENSDART00000164361
|
gcgra
|
glucagon receptor a |
| chr22_-_2870591 | 1.32 |
ENSDART00000063533
|
aqp12
|
aquaporin 12 |
| chr5_-_70939686 | 1.32 |
|
|
|
| chr19_-_5416317 | 1.31 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr2_-_47578695 | 1.30 |
ENSDART00000014350
ENSDART00000038828 |
pax3a
|
paired box 3a |
| chr2_+_33399405 | 1.30 |
ENSDART00000137207
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr13_+_14873339 | 1.30 |
ENSDART00000057810
|
emx1
|
empty spiracles homeobox 1 |
| chr18_+_45893199 | 1.29 |
ENSDART00000158246
|
dvl3b
|
dishevelled segment polarity protein 3b |
| chr3_-_23444371 | 1.29 |
ENSDART00000087726
|
igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr17_+_381715 | 1.29 |
ENSDART00000162898
|
si:rp71-62i8.1
|
si:rp71-62i8.1 |
| chr24_-_12794672 | 1.28 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr16_+_46145286 | 1.28 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr5_-_36647498 | 1.28 |
ENSDART00000112312
|
lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr12_+_35018663 | 1.28 |
ENSDART00000085774
|
si:ch73-127m5.1
|
si:ch73-127m5.1 |
| chr14_+_35906366 | 1.27 |
ENSDART00000105602
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr19_-_30816780 | 1.27 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr5_+_3172101 | 1.27 |
ENSDART00000080486
|
ywhag1
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 |
| chr7_-_39170016 | 1.26 |
ENSDART00000161191
|
BX842701.1
|
ENSDARG00000101872 |
| chr23_+_27985224 | 1.24 |
ENSDART00000171859
|
ENSDARG00000100606
|
ENSDARG00000100606 |
| chr5_-_25866099 | 1.23 |
ENSDART00000144035
|
arvcfb
|
armadillo repeat gene deleted in velocardiofacial syndrome b |
| chr6_+_12618821 | 1.23 |
ENSDART00000156341
|
fam117ba
|
family with sequence similarity 117, member Ba |
| chr14_-_46359062 | 1.23 |
ENSDART00000090844
|
zgc:153018
|
zgc:153018 |
| chr5_+_40722565 | 1.22 |
ENSDART00000097546
|
arid3c
|
AT rich interactive domain 3C (BRIGHT-like) |
| chr19_+_712139 | 1.20 |
ENSDART00000093281
|
fhod3a
|
formin homology 2 domain containing 3a |
| chr18_+_20504980 | 1.20 |
ENSDART00000060295
|
rapsn
|
receptor-associated protein of the synapse, 43kD |
| chr9_-_22488422 | 1.19 |
ENSDART00000124600
|
crygm2d21
|
crystallin, gamma M2d21 |
| chr2_-_31319359 | 1.19 |
ENSDART00000060812
|
adcyap1b
|
adenylate cyclase activating polypeptide 1b |
| chr6_+_36965023 | 1.19 |
ENSDART00000028895
|
negr1
|
neuronal growth regulator 1 |
| chr25_+_23494481 | 1.18 |
ENSDART00000089199
|
cpt1ab
|
carnitine palmitoyltransferase 1Ab (liver) |
| chr16_+_21436660 | 1.17 |
ENSDART00000145886
|
osbpl3b
|
oxysterol binding protein-like 3b |
| chr10_-_38206060 | 1.17 |
ENSDART00000174882
|
FP016248.1
|
ENSDARG00000107804 |
| chr9_+_42112576 | 1.16 |
ENSDART00000130434
ENSDART00000007058 |
col18a1a
|
collagen type XVIII alpha 1 chain a |
| chr4_+_1750689 | 1.16 |
ENSDART00000146779
|
slc38a2
|
solute carrier family 38, member 2 |
| chr18_-_48498261 | 1.15 |
ENSDART00000146346
|
kcnj1a.6
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 6 |
| chr3_-_18426055 | 1.15 |
ENSDART00000122968
|
aqp8b
|
aquaporin 8b |
| chr14_+_38375351 | 1.15 |
ENSDART00000175744
|
BX088707.3
|
ENSDARG00000107351 |
| chr7_+_36267647 | 1.14 |
ENSDART00000173653
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr4_-_16344954 | 1.14 |
ENSDART00000079523
|
epyc
|
epiphycan |
| chr20_+_40247918 | 1.14 |
ENSDART00000121818
|
trdn
|
triadin |
| chr10_+_13251463 | 1.13 |
ENSDART00000000887
|
rassf6
|
Ras association (RalGDS/AF-6) domain family 6 |
| chr15_+_27431469 | 1.12 |
ENSDART00000122101
|
tbx2b
|
T-box 2b |
| chr6_-_586251 | 1.12 |
ENSDART00000148867
ENSDART00000149414 ENSDART00000149248 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr7_+_25861386 | 1.11 |
ENSDART00000129834
|
nat16
|
N-acetyltransferase 16 |
| chr24_+_16249188 | 1.11 |
ENSDART00000164516
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr17_+_17935213 | 1.11 |
ENSDART00000104999
|
ccdc85ca
|
coiled-coil domain containing 85C, a |
| chr5_+_15167637 | 1.11 |
ENSDART00000127015
|
srrm4
|
serine/arginine repetitive matrix 4 |
| chr15_-_2969420 | 1.10 |
|
|
|
| chr18_+_9413588 | 1.10 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr1_-_48853800 | 1.09 |
ENSDART00000137357
|
zgc:175214
|
zgc:175214 |
| chr16_+_1338291 | 1.09 |
ENSDART00000149299
|
cers2b
|
ceramide synthase 2b |
| chr21_-_3301751 | 1.09 |
ENSDART00000009740
|
smad7
|
SMAD family member 7 |
| chr19_-_24859287 | 1.08 |
ENSDART00000163763
|
thbs3b
|
thrombospondin 3b |
| chr22_+_16511562 | 1.08 |
|
|
|
| chr16_+_53238110 | 1.07 |
ENSDART00000102170
|
CABZ01053976.1
|
ENSDARG00000069929 |
| chr7_-_33994315 | 1.07 |
ENSDART00000173596
|
si:ch211-98n17.5
|
si:ch211-98n17.5 |
| chr17_+_16067769 | 1.07 |
ENSDART00000155292
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr13_+_22350043 | 1.06 |
ENSDART00000136863
|
ldb3a
|
LIM domain binding 3a |
| chr1_+_16681778 | 1.05 |
|
|
|
| chr15_-_2689005 | 1.05 |
ENSDART00000063325
|
cldnf
|
claudin f |
| chr23_-_31446156 | 1.05 |
ENSDART00000053367
|
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| KN150683v1_+_13359 | 1.04 |
|
|
|
| chr16_-_42968807 | 1.04 |
ENSDART00000154757
|
txnipb
|
thioredoxin interacting protein b |
| chr2_-_30675594 | 1.03 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr5_+_69383382 | 1.03 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr12_-_457997 | 1.02 |
ENSDART00000143232
|
dhrs7cb
|
dehydrogenase/reductase (SDR family) member 7Cb |
| chr20_-_11179880 | 1.02 |
ENSDART00000152246
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr6_+_3549861 | 1.02 |
ENSDART00000170781
|
phospho2
|
phosphatase, orphan 2 |
| chr6_+_44817077 | 1.01 |
ENSDART00000169713
|
chl1b
|
cell adhesion molecule L1-like b |
| chr7_-_2367001 | 1.01 |
ENSDART00000174011
|
si:cabz01007812.1
|
si:cabz01007812.1 |
| chr18_+_36650892 | 1.00 |
ENSDART00000098980
|
znf296
|
zinc finger protein 296 |
| chr6_-_49900124 | 1.00 |
ENSDART00000150204
|
atp5e
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
| chr8_-_11512545 | 0.99 |
ENSDART00000133932
|
si:ch211-248e11.2
|
si:ch211-248e11.2 |
| chr16_-_1479139 | 0.99 |
ENSDART00000036348
|
sim1a
|
single-minded family bHLH transcription factor 1a |
| chr19_-_9793494 | 0.99 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr20_-_40217185 | 0.99 |
|
|
|
| chr9_-_711269 | 0.98 |
ENSDART00000144625
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr23_+_18796386 | 0.98 |
ENSDART00000137438
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr1_-_11591684 | 0.97 |
ENSDART00000003825
|
cplx2l
|
complexin 2, like |
| chr8_+_24725855 | 0.97 |
ENSDART00000078656
|
slc16a4
|
solute carrier family 16, member 4 |
| chr4_-_25282256 | 0.97 |
ENSDART00000066936
|
tmem110l
|
transmembrane protein 110, like |
| chr25_-_22890657 | 0.97 |
|
|
|
| chr2_+_52473102 | 0.96 |
ENSDART00000146418
|
shda
|
Src homology 2 domain containing transforming protein D, a |
| chr9_-_22377136 | 0.96 |
ENSDART00000110933
|
CR847505.1
|
ENSDARG00000076693 |
| chr24_-_23829288 | 0.96 |
ENSDART00000080510
ENSDART00000135242 |
ENSDARG00000019332
|
ENSDARG00000019332 |
| chr5_-_54150484 | 0.96 |
ENSDART00000058470
|
pik3r1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr5_-_63031745 | 0.96 |
|
|
|
| chr6_+_48138737 | 0.95 |
|
|
|
| chr10_+_34372205 | 0.95 |
ENSDART00000110121
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr3_-_13942505 | 0.95 |
|
|
|
| chr10_-_38206132 | 0.95 |
ENSDART00000174882
|
FP016248.1
|
ENSDARG00000107804 |
| chr8_-_2447758 | 0.95 |
ENSDART00000101125
|
rpl6
|
ribosomal protein L6 |
| chr9_+_21725007 | 0.95 |
ENSDART00000045093
|
arhgef7a
|
Rho guanine nucleotide exchange factor (GEF) 7a |
| chr14_-_6822151 | 0.95 |
ENSDART00000171311
|
si:ch73-43g23.1
|
si:ch73-43g23.1 |
| chr23_-_3760604 | 0.95 |
ENSDART00000143731
|
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr18_-_11439124 | 0.95 |
|
|
|
| chr11_-_33925792 | 0.95 |
ENSDART00000172978
|
atp13a3
|
ATPase type 13A3 |
| chr8_-_49443058 | 0.94 |
ENSDART00000011453
|
sypb
|
synaptophysin b |
| chr8_+_23192085 | 0.94 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr25_-_33920256 | 0.94 |
|
|
|
| chr14_-_48951428 | 0.93 |
ENSDART00000157785
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr7_+_24770873 | 0.92 |
ENSDART00000165314
|
AL954715.1
|
ENSDARG00000103682 |
| chr8_+_48624273 | 0.92 |
ENSDART00000121432
|
nppa
|
natriuretic peptide A |
| chr7_+_28862183 | 0.92 |
ENSDART00000052346
|
gnao1b
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, b |
| chr25_+_35901081 | 0.92 |
ENSDART00000042271
ENSDART00000158027 |
irx3b
|
iroquois homeobox 3b |
| chr5_-_43896815 | 0.92 |
ENSDART00000110076
|
gas1a
|
growth arrest-specific 1a |
| chr11_-_6004509 | 0.92 |
ENSDART00000108628
|
ano8b
|
anoctamin 8b |
| chr5_-_31301280 | 0.92 |
ENSDART00000141446
|
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr6_+_1895734 | 0.91 |
ENSDART00000109679
|
quo
|
quattro |
| chr20_+_3095763 | 0.91 |
ENSDART00000133435
|
CEP170B
|
centrosomal protein 170B |
| chr16_-_17439735 | 0.91 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr6_-_39608334 | 0.91 |
ENSDART00000179059
|
dip2bb
|
disco-interacting protein 2 homolog Bb |
| chr13_-_39821399 | 0.91 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.3 | GO:1905178 | regulation of cardiac muscle tissue regeneration(GO:1905178) |
| 1.2 | 4.8 | GO:0071549 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) glycerol biosynthetic process from pyruvate(GO:0046327) cellular response to dexamethasone stimulus(GO:0071549) |
| 1.1 | 4.5 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.9 | 2.8 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) |
| 0.7 | 2.1 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) positive regulation of muscle contraction(GO:0045933) |
| 0.7 | 3.3 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.6 | 3.6 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.6 | 1.8 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.5 | 1.0 | GO:0071331 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.5 | 1.4 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.5 | 1.4 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.4 | 1.7 | GO:0015859 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.4 | 2.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.4 | 2.3 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.3 | 1.4 | GO:0048664 | auditory receptor cell fate commitment(GO:0009912) neuron fate determination(GO:0048664) inner ear receptor cell fate commitment(GO:0060120) |
| 0.3 | 1.0 | GO:0035776 | pronephric proximal tubule development(GO:0035776) |
| 0.3 | 1.3 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.3 | 0.9 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.3 | 2.0 | GO:0072132 | mesenchyme morphogenesis(GO:0072132) |
| 0.3 | 1.4 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.3 | 3.7 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.3 | 0.8 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.3 | 2.2 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.3 | 1.5 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.3 | 1.3 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.2 | 1.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 0.9 | GO:0003085 | negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.2 | 3.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 0.6 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.2 | 1.3 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.2 | 1.3 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.2 | 0.8 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.2 | 1.3 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.2 | 0.7 | GO:0061036 | positive regulation of chondrocyte differentiation(GO:0032332) positive regulation of cartilage development(GO:0061036) |
| 0.2 | 0.5 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of BMP from receptor via BMP binding(GO:0038098) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.2 | 12.1 | GO:0007601 | visual perception(GO:0007601) |
| 0.2 | 1.9 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.2 | 1.7 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.2 | 1.0 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) |
| 0.2 | 0.5 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.2 | 0.5 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 | 1.2 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.2 | 1.3 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.2 | 0.8 | GO:0010991 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.2 | 0.6 | GO:0072314 | visceral serous pericardium development(GO:0061032) glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.2 | 0.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 0.5 | GO:0052805 | imidazole-containing compound metabolic process(GO:0052803) imidazole-containing compound catabolic process(GO:0052805) |
| 0.1 | 0.4 | GO:0061114 | pancreas morphogenesis(GO:0061113) branching involved in pancreas morphogenesis(GO:0061114) pancreatic bud formation(GO:0061130) pancreas field specification(GO:0061131) |
| 0.1 | 0.4 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.1 | 0.4 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.6 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 0.7 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 1.1 | GO:0042551 | neuron maturation(GO:0042551) |
| 0.1 | 0.4 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.7 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.1 | 1.7 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 2.1 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 1.4 | GO:0046037 | GMP biosynthetic process(GO:0006177) GTP biosynthetic process(GO:0006183) GMP metabolic process(GO:0046037) |
| 0.1 | 0.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 1.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.6 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.9 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 0.7 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.1 | 0.8 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.1 | 0.2 | GO:0045117 | azole transport(GO:0045117) |
| 0.1 | 0.9 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 1.6 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 1.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 0.8 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.1 | 0.3 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.8 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 | 0.3 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.9 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 1.9 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 1.5 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.1 | 0.3 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.1 | 1.0 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.3 | GO:0048070 | regulation of developmental pigmentation(GO:0048070) |
| 0.1 | 1.0 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 0.3 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 0.7 | GO:2000251 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 1.7 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 1.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.5 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 1.3 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 0.5 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 1.5 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.1 | 0.4 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 1.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 1.8 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 1.8 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 0.8 | GO:0061099 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 5.4 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 0.6 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 0.4 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 4.1 | GO:0071560 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.1 | 1.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 1.7 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.1 | 0.4 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.8 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.4 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.8 | GO:0050773 | regulation of dendrite development(GO:0050773) |
| 0.0 | 0.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.2 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 1.0 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 0.4 | GO:0006188 | IMP biosynthetic process(GO:0006188) |
| 0.0 | 0.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 3.3 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.6 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 2.0 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.9 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 1.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.9 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.0 | 0.4 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.5 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.9 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 1.4 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.3 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 1.4 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 1.6 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.0 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.2 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 1.5 | GO:0001817 | regulation of cytokine production(GO:0001817) |
| 0.0 | 1.4 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 1.3 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.6 | GO:0046460 | triglyceride biosynthetic process(GO:0019432) neutral lipid biosynthetic process(GO:0046460) acylglycerol biosynthetic process(GO:0046463) |
| 0.0 | 0.1 | GO:0043627 | response to estrogen(GO:0043627) |
| 0.0 | 1.3 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.7 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 3.0 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.4 | GO:1902667 | regulation of axon guidance(GO:1902667) |
| 0.0 | 1.2 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.8 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 1.2 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.4 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.5 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 1.1 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.1 | GO:0044209 | purine nucleotide salvage(GO:0032261) AMP salvage(GO:0044209) |
| 0.0 | 1.4 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.1 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.3 | GO:0043409 | negative regulation of MAPK cascade(GO:0043409) |
| 0.0 | 0.7 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.1 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.2 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.2 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 2.7 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.5 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 0.3 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 0.3 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.0 | 0.2 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 1.0 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0036010 | protein localization to endosome(GO:0036010) protein localization to early endosome(GO:1902946) |
| 0.0 | 0.4 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.9 | GO:0031430 | M band(GO:0031430) |
| 0.4 | 2.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.3 | 3.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.3 | 5.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 2.2 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.2 | 1.8 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 2.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.2 | 2.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 1.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 1.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 2.0 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 5.0 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 1.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.4 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 1.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 6.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 2.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 2.2 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 1.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 2.3 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 0.2 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 1.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.6 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 1.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.7 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 24.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.3 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 3.9 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0030125 | clathrin vesicle coat(GO:0030125) clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 5.9 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.1 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.0 | 1.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.0 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.6 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.2 | GO:0015629 | actin cytoskeleton(GO:0015629) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.8 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.7 | 7.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.5 | 2.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.5 | 1.9 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.5 | 1.4 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.4 | 1.7 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.4 | 1.7 | GO:0033971 | hydroxyisourate hydrolase activity(GO:0033971) |
| 0.4 | 12.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.4 | 2.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.4 | 1.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.4 | 3.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 1.4 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.3 | 0.7 | GO:2001070 | starch binding(GO:2001070) |
| 0.3 | 2.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.3 | 2.2 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.3 | 1.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.2 | 0.7 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 3.3 | GO:0015245 | long-chain fatty acid transporter activity(GO:0005324) fatty acid transporter activity(GO:0015245) |
| 0.2 | 1.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 1.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.2 | 0.7 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.2 | 1.8 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.2 | 0.6 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.2 | 1.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 0.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 1.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.2 | 1.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 2.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 1.6 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 1.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 2.2 | GO:0043236 | laminin binding(GO:0043236) |
| 0.2 | 0.7 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 0.8 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 1.8 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 0.8 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 1.8 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 1.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.8 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 1.8 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 3.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 1.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 1.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.4 | GO:0070699 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.1 | 0.2 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.1 | 2.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 1.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.9 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.4 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.7 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.5 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.9 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.4 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.5 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 1.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.7 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.6 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.1 | 4.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 0.4 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.1 | 0.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.3 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.4 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.1 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.7 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.1 | 0.5 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.4 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 1.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.8 | GO:0016722 | oxidoreductase activity, oxidizing metal ions(GO:0016722) |
| 0.1 | 2.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 1.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.4 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 1.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 3.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.1 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.1 | 0.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 1.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.5 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 2.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 11.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.9 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 1.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 2.3 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0019201 | nucleotide kinase activity(GO:0019201) |
| 0.0 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.5 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.0 | 1.0 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.6 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.8 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 1.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 1.4 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 3.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 1.0 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 12.1 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.2 | 2.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.2 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 2.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 5.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.9 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 2.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 2.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.3 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.3 | 1.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 3.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 4.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.2 | 1.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 1.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.8 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 0.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.8 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.6 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 2.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.7 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 3.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 0.7 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.7 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.8 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 2.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |