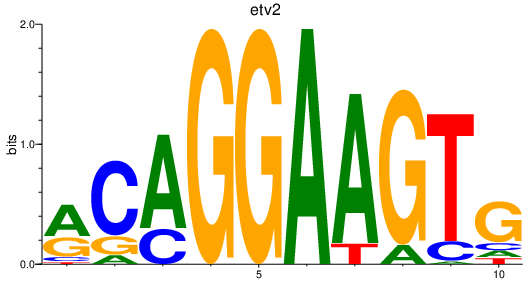
Project
DANIO-CODE
Navigation
Downloads
Results for etv2_erg_fev+fli1a
Z-value: 1.71
Transcription factors associated with etv2_erg_fev+fli1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
etv2
|
ENSDARG00000053868 | ETS variant transcription factor 2 |
|
erg
|
ENSDARG00000077304 | ETS transcription factor ERG |
|
fev
|
ENSDARG00000009242 | FEV transcription factor, ETS family member |
|
fli1a
|
ENSDARG00000054632 | Fli-1 proto-oncogene, ETS transcription factor a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| fli1a | dr10_dc_chr18_+_48433986_48434041 | -0.90 | 2.6e-06 | Click! |
| etv2 | dr10_dc_chr16_-_42063851_42063895 | -0.81 | 1.5e-04 | Click! |
Activity profile of etv2_erg_fev+fli1a motif
Sorted Z-values of etv2_erg_fev+fli1a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of etv2_erg_fev+fli1a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_4264663 | 7.30 |
ENSDART00000152521
|
ca15b
|
carbonic anhydrase XVb |
| chr12_-_42212994 | 7.06 |
ENSDART00000171075
|
zgc:111868
|
zgc:111868 |
| chr6_-_10492276 | 5.18 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr18_-_22745677 | 4.64 |
ENSDART00000023721
|
nudt21
|
nudix hydrolase 21 |
| chr17_+_23463072 | 4.34 |
|
|
|
| chr22_-_17627900 | 4.32 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr6_-_10492123 | 4.14 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| KN150623v1_+_258 | 4.10 |
|
|
|
| chr24_+_31522539 | 3.98 |
ENSDART00000126380
|
cnbd1
|
cyclic nucleotide binding domain containing 1 |
| chr19_-_20819101 | 3.94 |
ENSDART00000137590
|
dazl
|
deleted in azoospermia-like |
| chr10_-_180603 | 3.92 |
|
|
|
| chr16_-_46426385 | 3.82 |
ENSDART00000141331
|
si:ch73-59c19.1
|
si:ch73-59c19.1 |
| chr13_-_37340209 | 3.80 |
|
|
|
| chr20_-_34126039 | 3.79 |
ENSDART00000033817
|
scyl3
|
SCY1-like, kinase-like 3 |
| chr14_-_880799 | 3.78 |
ENSDART00000031992
|
rgs14a
|
regulator of G protein signaling 14a |
| chr24_-_9857510 | 3.76 |
ENSDART00000136274
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr18_-_3455211 | 3.64 |
ENSDART00000163762
|
dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr25_+_4888647 | 3.57 |
ENSDART00000165933
ENSDART00000159537 |
hdac10
|
histone deacetylase 10 |
| chr2_+_28199458 | 3.53 |
ENSDART00000150330
|
buc
|
bucky ball |
| chr25_-_9889107 | 3.50 |
ENSDART00000137407
|
AL929493.1
|
ENSDARG00000093575 |
| chr22_-_17627831 | 3.50 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr15_+_21776031 | 3.33 |
ENSDART00000136151
|
NKAPD1
|
zgc:162339 |
| KN150663v1_-_3381 | 3.22 |
|
|
|
| chr18_-_11626694 | 3.14 |
ENSDART00000098565
|
CRACR2A
|
calcium release activated channel regulator 2A |
| chr5_-_31796376 | 3.11 |
ENSDART00000142095
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr8_+_52456064 | 3.08 |
ENSDART00000012758
|
zgc:77112
|
zgc:77112 |
| chr3_+_32285237 | 3.08 |
ENSDART00000157324
|
prrg2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr5_-_15971338 | 3.03 |
ENSDART00000110437
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr8_+_8605240 | 3.02 |
ENSDART00000075624
|
usp11
|
ubiquitin specific peptidase 11 |
| chr14_+_32578103 | 3.00 |
ENSDART00000105721
|
lnx2b
|
ligand of numb-protein X 2b |
| chr19_+_43928043 | 2.99 |
ENSDART00000113031
|
wasf2
|
WAS protein family, member 2 |
| chr6_+_49772891 | 2.93 |
ENSDART00000134207
|
ctsz
|
cathepsin Z |
| chr14_+_31596468 | 2.92 |
ENSDART00000173259
|
BX005454.1
|
ENSDARG00000100646 |
| chr23_-_16755868 | 2.91 |
ENSDART00000020810
|
sdcbp2
|
syndecan binding protein (syntenin) 2 |
| chr12_-_13692190 | 2.85 |
ENSDART00000152370
|
foxh1
|
forkhead box H1 |
| chr9_+_8387050 | 2.83 |
ENSDART00000136847
|
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr7_+_28824986 | 2.80 |
ENSDART00000052349
|
ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr3_+_42395984 | 2.80 |
ENSDART00000162096
|
micall2a
|
mical-like 2a |
| chr22_+_4035577 | 2.72 |
ENSDART00000170620
|
ctxn1
|
cortexin 1 |
| chr20_+_16740320 | 2.71 |
ENSDART00000152359
|
tmem30ab
|
transmembrane protein 30Ab |
| chr16_-_34240818 | 2.69 |
ENSDART00000054026
|
rcc1
|
regulator of chromosome condensation 1 |
| chr14_+_32578253 | 2.69 |
ENSDART00000105721
|
lnx2b
|
ligand of numb-protein X 2b |
| chr20_+_27188382 | 2.65 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr24_-_38195458 | 2.63 |
ENSDART00000056381
|
crp2
|
C-reactive protein 2 |
| chr1_+_157840 | 2.62 |
ENSDART00000152205
ENSDART00000160843 |
cul4a
|
cullin 4A |
| chr7_-_47990610 | 2.62 |
ENSDART00000147968
|
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr25_-_3619590 | 2.61 |
ENSDART00000037973
|
morc2
|
MORC family CW-type zinc finger 2 |
| chr22_-_20377970 | 2.61 |
ENSDART00000010048
|
map2k2a
|
mitogen-activated protein kinase kinase 2a |
| chr5_-_41260390 | 2.61 |
ENSDART00000140154
|
si:dkey-65b12.12
|
si:dkey-65b12.12 |
| chr7_-_20556760 | 2.61 |
ENSDART00000143509
|
dnajc3b
|
DnaJ (Hsp40) homolog, subfamily C, member 3b |
| chr16_+_47283253 | 2.54 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr14_-_47210912 | 2.51 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr13_-_17991273 | 2.45 |
ENSDART00000128748
|
fam21c
|
family with sequence similarity 21, member C |
| chr3_+_32700459 | 2.41 |
ENSDART00000132679
ENSDART00000035759 |
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr1_-_51870424 | 2.38 |
ENSDART00000135636
|
acy3.1
|
aspartoacylase (aminocyclase) 3, tandem duplicate 1 |
| chr20_+_24047842 | 2.38 |
ENSDART00000128616
ENSDART00000144195 |
casp8ap2
|
caspase 8 associated protein 2 |
| chr5_-_31796734 | 2.38 |
ENSDART00000142095
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr4_+_12967786 | 2.36 |
ENSDART00000113357
|
vhll
|
von Hippel-Lindau tumor suppressor like |
| chr5_-_38570658 | 2.34 |
ENSDART00000020808
|
paqr3a
|
progestin and adipoQ receptor family member IIIa |
| chr20_+_2100836 | 2.33 |
|
|
|
| chr8_+_36522231 | 2.31 |
ENSDART00000126687
|
sf3a1
|
splicing factor 3a, subunit 1 |
| chr11_+_24076334 | 2.31 |
ENSDART00000017599
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr18_+_15303434 | 2.30 |
ENSDART00000099777
ENSDART00000170246 |
si:dkey-103i16.6
|
si:dkey-103i16.6 |
| chr20_+_37592206 | 2.29 |
ENSDART00000153092
|
BX088686.1
|
ENSDARG00000096774 |
| chr13_+_22973764 | 2.26 |
ENSDART00000110266
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr10_-_180681 | 2.24 |
|
|
|
| chr12_-_2834771 | 2.23 |
ENSDART00000114854
ENSDART00000163759 |
zdhhc16b
|
zinc finger, DHHC-type containing 16b |
| chr14_+_24543399 | 2.23 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr11_+_36088518 | 2.22 |
|
|
|
| chr23_-_37010705 | 2.21 |
ENSDART00000134461
|
zgc:193690
|
zgc:193690 |
| chr9_-_55878791 | 2.21 |
|
|
|
| chr3_+_31530419 | 2.19 |
ENSDART00000113441
|
mylk5
|
myosin, light chain kinase 5 |
| chr14_-_27040866 | 2.19 |
ENSDART00000173053
|
CU062454.1
|
ENSDARG00000105327 |
| chr21_+_28710341 | 2.19 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr24_-_38195383 | 2.18 |
ENSDART00000056381
|
crp2
|
C-reactive protein 2 |
| chr6_-_39633603 | 2.18 |
ENSDART00000104042
|
atf7b
|
activating transcription factor 7b |
| chr5_+_9579430 | 2.18 |
ENSDART00000109236
|
ENSDARG00000075416
|
ENSDARG00000075416 |
| chr5_-_23741221 | 2.18 |
ENSDART00000042481
|
phf23a
|
PHD finger protein 23a |
| chr16_-_25318413 | 2.18 |
ENSDART00000058943
|
ENSDARG00000040280
|
ENSDARG00000040280 |
| chr5_-_56641061 | 2.15 |
ENSDART00000050957
|
fer
|
fer (fps/fes related) tyrosine kinase |
| chr15_+_28173644 | 2.15 |
ENSDART00000041707
|
unc119a
|
unc-119 homolog a (C. elegans) |
| KN150307v1_+_6450 | 2.14 |
ENSDART00000159959
|
CABZ01038161.1
|
ENSDARG00000098784 |
| chr13_-_24265471 | 2.14 |
ENSDART00000016211
|
tbp
|
TATA box binding protein |
| chr16_+_38251653 | 2.11 |
ENSDART00000044971
|
myo1eb
|
myosin IE, b |
| chr7_+_28825033 | 2.11 |
ENSDART00000052349
|
ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr19_+_26756510 | 2.11 |
ENSDART00000013497
|
mylipa
|
myosin regulatory light chain interacting protein a |
| chr6_+_13766964 | 2.11 |
ENSDART00000136006
ENSDART00000009382 |
dnpep
|
aspartyl aminopeptidase |
| chr3_-_32457708 | 2.10 |
|
|
|
| chr8_-_49227286 | 2.09 |
|
|
|
| chr8_-_49227232 | 2.09 |
|
|
|
| chr17_-_33764220 | 2.08 |
ENSDART00000043651
|
dnal1
|
dynein, axonemal, light chain 1 |
| chr5_-_23092592 | 2.08 |
ENSDART00000024815
|
fam76b
|
family with sequence similarity 76, member B |
| chr24_-_9874069 | 2.08 |
ENSDART00000176344
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr14_+_31734245 | 2.08 |
ENSDART00000172754
ENSDART00000163876 |
BX511142.1
|
ENSDARG00000099086 |
| chr24_+_43010 | 2.07 |
ENSDART00000122785
|
zgc:152808
|
zgc:152808 |
| chr11_+_30400284 | 2.05 |
ENSDART00000169833
|
gb:eh507706
|
expressed sequence EH507706 |
| chr23_+_17999913 | 2.05 |
ENSDART00000012540
|
chia.4
|
chitinase, acidic.4 |
| chr17_+_7377230 | 2.04 |
ENSDART00000157123
|
shprh
|
SNF2 histone linker PHD RING helicase |
| chr15_+_17407036 | 2.04 |
ENSDART00000018461
|
vmp1
|
vacuole membrane protein 1 |
| chr10_-_10907273 | 2.03 |
|
|
|
| chr18_+_48188699 | 2.02 |
|
|
|
| chr12_+_48590478 | 2.02 |
|
|
|
| chr7_+_22524917 | 2.02 |
ENSDART00000112169
|
rbm4.2
|
RNA binding motif protein 4.2 |
| KN149841v1_+_6481 | 2.01 |
ENSDART00000179534
|
CABZ01103873.1
|
ENSDARG00000108002 |
| chr12_-_4264610 | 2.01 |
ENSDART00000152377
|
ca15b
|
carbonic anhydrase XVb |
| chr1_+_58608185 | 2.01 |
ENSDART00000161872
ENSDART00000160658 |
parn
|
poly(A)-specific ribonuclease (deadenylation nuclease) |
| chr19_+_43984165 | 2.00 |
ENSDART00000138404
|
si:ch211-199g17.2
|
si:ch211-199g17.2 |
| chr15_+_44909293 | 1.99 |
ENSDART00000111373
|
pdgfd
|
platelet derived growth factor d |
| chr8_+_25072241 | 1.99 |
ENSDART00000143922
|
atxn7l2b
|
ataxin 7-like 2b |
| chr16_+_14075878 | 1.98 |
ENSDART00000059926
|
zgc:162509
|
zgc:162509 |
| chr7_+_24257251 | 1.95 |
ENSDART00000136473
|
ENSDARG00000079281
|
ENSDARG00000079281 |
| chr1_+_18909404 | 1.93 |
ENSDART00000088933
ENSDART00000141579 |
fbxo10
|
F-box protein 10 |
| chr25_-_19889225 | 1.92 |
|
|
|
| chr24_+_16575167 | 1.92 |
|
|
|
| chr1_+_50547385 | 1.91 |
ENSDART00000132141
|
btbd3a
|
BTB (POZ) domain containing 3a |
| chr16_+_53366182 | 1.91 |
ENSDART00000162926
|
si:ch211-269k10.5
|
si:ch211-269k10.5 |
| chr22_+_23260371 | 1.91 |
|
|
|
| chr9_-_32347822 | 1.90 |
ENSDART00000037182
|
ankrd44
|
ankyrin repeat domain 44 |
| chr10_-_25725495 | 1.90 |
ENSDART00000177220
|
AL837524.2
|
ENSDARG00000108650 |
| chr14_-_26167494 | 1.90 |
ENSDART00000143454
ENSDART00000111860 |
syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr2_+_30199024 | 1.89 |
ENSDART00000004903
|
rdh10b
|
retinol dehydrogenase 10b |
| chr6_+_12810986 | 1.89 |
ENSDART00000104757
ENSDART00000165896 |
casp8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr19_-_30245434 | 1.88 |
ENSDART00000167378
|
eif3i
|
eukaryotic translation initiation factor 3, subunit I |
| chr13_-_9034919 | 1.87 |
ENSDART00000135373
|
si:dkey-33c12.4
|
si:dkey-33c12.4 |
| chr2_+_12547264 | 1.87 |
ENSDART00000169724
|
zgc:66475
|
zgc:66475 |
| chr5_+_22541236 | 1.87 |
ENSDART00000171719
|
atrxl
|
alpha thalassemia/mental retardation syndrome X-linked, like |
| chr25_-_21684852 | 1.87 |
ENSDART00000089616
|
bmt2
|
base methyltransferase of 25S rRNA 2 homolog |
| chr9_-_32348003 | 1.86 |
ENSDART00000037182
|
ankrd44
|
ankyrin repeat domain 44 |
| chr16_+_43464824 | 1.86 |
ENSDART00000032778
|
rnf144b
|
ring finger protein 144B |
| chr25_-_25480657 | 1.86 |
ENSDART00000067138
|
hic1l
|
hypermethylated in cancer 1 like |
| chr18_+_9755131 | 1.86 |
|
|
|
| chr23_+_30803518 | 1.85 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr6_-_19556029 | 1.85 |
ENSDART00000136019
|
ppp1r12c
|
protein phosphatase 1, regulatory subunit 12C |
| chr16_-_26654610 | 1.85 |
ENSDART00000134448
|
l3mbtl1b
|
l(3)mbt-like 1b (Drosophila) |
| chr2_+_37224986 | 1.84 |
ENSDART00000138952
|
apoda.2
|
apolipoprotein Da, duplicate 2 |
| chr5_-_26195227 | 1.84 |
ENSDART00000146124
|
si:ch211-102c2.7
|
si:ch211-102c2.7 |
| chr17_-_7575628 | 1.83 |
ENSDART00000064657
|
stx11a
|
syntaxin 11a |
| chr14_-_20940726 | 1.83 |
ENSDART00000129743
|
si:ch211-175m2.5
|
si:ch211-175m2.5 |
| chr17_+_44327269 | 1.83 |
ENSDART00000045882
|
ap5m1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr18_+_20045544 | 1.81 |
|
|
|
| chr8_+_45326435 | 1.81 |
ENSDART00000134161
|
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr21_+_19040595 | 1.80 |
ENSDART00000145969
|
BX000991.1
|
ENSDARG00000092282 |
| chr16_+_47283374 | 1.80 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr6_-_10492724 | 1.80 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr7_-_68962286 | 1.79 |
|
|
|
| chr11_+_24076576 | 1.79 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr16_+_25344257 | 1.78 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| chr23_-_42918086 | 1.78 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr22_+_20686657 | 1.78 |
|
|
|
| chr15_+_38397715 | 1.78 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr5_+_71100066 | 1.77 |
ENSDART00000115182
|
nup214
|
nucleoporin 214 |
| chr23_-_32267833 | 1.77 |
|
|
|
| chr25_+_27301180 | 1.76 |
ENSDART00000149456
|
wasla
|
Wiskott-Aldrich syndrome-like a |
| chr20_-_49067167 | 1.76 |
ENSDART00000163071
ENSDART00000170617 |
xrn2
|
5'-3' exoribonuclease 2 |
| chr6_+_40556891 | 1.76 |
ENSDART00000155928
|
ddi2
|
DNA-damage inducible protein 2 |
| chr25_+_32716795 | 1.75 |
ENSDART00000131098
|
ENSDARG00000087402
|
ENSDARG00000087402 |
| chr9_+_8919663 | 1.73 |
ENSDART00000134954
|
carkd
|
carbohydrate kinase domain containing |
| chr6_-_44164161 | 1.73 |
ENSDART00000035513
|
shq1
|
SHQ1, H/ACA ribonucleoprotein assembly factor |
| chr15_+_30246345 | 1.72 |
ENSDART00000100214
|
nufip2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr10_-_36848872 | 1.72 |
|
|
|
| KN150065v1_-_622 | 1.72 |
|
|
|
| chr5_-_38571198 | 1.72 |
ENSDART00000020808
|
paqr3a
|
progestin and adipoQ receptor family member IIIa |
| chr24_+_21030141 | 1.71 |
ENSDART00000154940
|
naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr13_-_33348231 | 1.71 |
ENSDART00000160520
|
mad2l1bp
|
MAD2L1 binding protein |
| chr8_-_22537326 | 1.71 |
ENSDART00000165640
|
porcnl
|
porcupine homolog like |
| chr17_-_44326992 | 1.71 |
ENSDART00000148786
|
exoc5
|
exocyst complex component 5 |
| chr18_-_3455306 | 1.70 |
ENSDART00000166841
ENSDART00000161197 |
dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr3_+_24510974 | 1.70 |
ENSDART00000148414
ENSDART00000055590 |
zgc:113411
|
zgc:113411 |
| chr3_+_42396136 | 1.70 |
ENSDART00000165708
|
micall2a
|
mical-like 2a |
| chr15_+_35081979 | 1.69 |
ENSDART00000131182
|
zgc:66024
|
zgc:66024 |
| chr13_-_22776767 | 1.69 |
ENSDART00000143097
|
rufy2
|
RUN and FYVE domain containing 2 |
| chr7_+_23881828 | 1.68 |
ENSDART00000076735
|
lrp10
|
low density lipoprotein receptor-related protein 10 |
| chr5_+_29791993 | 1.68 |
ENSDART00000086765
|
stk36
|
serine/threonine kinase 36 (fused homolog, Drosophila) |
| chr22_+_23260413 | 1.68 |
|
|
|
| chr5_-_56641212 | 1.68 |
ENSDART00000050957
|
fer
|
fer (fps/fes related) tyrosine kinase |
| chr15_-_28284662 | 1.68 |
|
|
|
| chr7_+_56787468 | 1.67 |
ENSDART00000024076
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr14_+_43357313 | 1.67 |
|
|
|
| chr8_-_47339228 | 1.66 |
ENSDART00000060853
|
pex10
|
peroxisomal biogenesis factor 10 |
| chr9_+_2756362 | 1.66 |
ENSDART00000001795
|
sp3a
|
sp3a transcription factor |
| chr13_-_36312975 | 1.66 |
ENSDART00000084867
|
dcaf5
|
ddb1 and cul4 associated factor 5 |
| chr19_-_6925090 | 1.66 |
ENSDART00000081568
|
tcf19l
|
transcription factor 19 (SC1), like |
| chr4_-_12979905 | 1.66 |
ENSDART00000150674
|
tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr20_-_22899048 | 1.65 |
ENSDART00000063609
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr13_+_15669924 | 1.65 |
ENSDART00000146234
|
apopt1
|
apoptogenic 1, mitochondrial |
| chr10_-_5135347 | 1.65 |
ENSDART00000138537
|
sec31a
|
SEC31 homolog A, COPII coat complex component |
| chr15_+_46093261 | 1.64 |
|
|
|
| chr5_+_68960543 | 1.64 |
ENSDART00000169013
|
arl6ip4
|
ADP-ribosylation factor-like 6 interacting protein 4 |
| chr24_-_36721857 | 1.63 |
|
|
|
| chr22_-_21021984 | 1.63 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr1_-_7962760 | 1.63 |
ENSDART00000103644
|
fbxl18
|
F-box and leucine-rich repeat protein 18 |
| chr17_-_7661725 | 1.63 |
ENSDART00000143870
|
rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr13_-_51743281 | 1.63 |
|
|
|
| chr19_+_16159980 | 1.62 |
ENSDART00000151169
|
gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr18_-_49291686 | 1.62 |
ENSDART00000174038
|
si:zfos-464b6.2
|
si:zfos-464b6.2 |
| chr1_-_44649941 | 1.62 |
ENSDART00000110390
|
ENSDARG00000025518
|
ENSDARG00000025518 |
| chr19_-_20819477 | 1.62 |
ENSDART00000151356
|
dazl
|
deleted in azoospermia-like |
| chr16_+_36794614 | 1.62 |
ENSDART00000139069
|
decr1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr6_+_31381325 | 1.61 |
ENSDART00000038990
|
jak1
|
Janus kinase 1 |
| chr2_-_57785550 | 1.61 |
ENSDART00000140060
|
si:dkeyp-68b7.7
|
si:dkeyp-68b7.7 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:2000975 | pro-B cell differentiation(GO:0002328) regulation of mRNA cleavage(GO:0031437) positive regulation of mRNA cleavage(GO:0031439) regulation of pro-B cell differentiation(GO:2000973) positive regulation of pro-B cell differentiation(GO:2000975) |
| 1.4 | 6.8 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 1.3 | 6.6 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 1.2 | 3.5 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 1.1 | 5.6 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 1.1 | 3.3 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 1.0 | 3.0 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 1.0 | 6.9 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.9 | 5.6 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.9 | 2.7 | GO:0000493 | box H/ACA snoRNP assembly(GO:0000493) |
| 0.8 | 2.5 | GO:0043903 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) |
| 0.8 | 4.9 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.7 | 8.4 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.7 | 2.8 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.7 | 3.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.7 | 2.6 | GO:0051150 | regulation of smooth muscle cell differentiation(GO:0051150) negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.6 | 14.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.6 | 2.4 | GO:2000040 | planar cell polarity pathway involved in axis elongation(GO:0003402) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) |
| 0.6 | 2.4 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.6 | 3.5 | GO:0030237 | female sex determination(GO:0030237) |
| 0.6 | 2.8 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.6 | 1.7 | GO:0001502 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.5 | 1.6 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.5 | 4.3 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.5 | 2.1 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.5 | 1.6 | GO:0070849 | response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.5 | 1.6 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.5 | 1.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.5 | 1.5 | GO:0060760 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.5 | 1.5 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.5 | 2.0 | GO:1902236 | regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902235) negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.5 | 0.5 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.5 | 3.0 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.5 | 3.3 | GO:0034087 | establishment of mitotic sister chromatid cohesion(GO:0034087) |
| 0.5 | 1.4 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.5 | 0.5 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.4 | 3.6 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.4 | 3.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.4 | 1.8 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.4 | 1.3 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine diphosphate metabolic process(GO:0042357) thiamine-containing compound metabolic process(GO:0042723) |
| 0.4 | 7.2 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.4 | 3.3 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.4 | 1.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.4 | 2.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.4 | 1.5 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.4 | 1.1 | GO:1903523 | negative regulation of heart contraction(GO:0045822) negative regulation of blood circulation(GO:1903523) |
| 0.4 | 5.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.4 | 1.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.4 | 4.4 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.4 | 6.9 | GO:0051351 | positive regulation of ligase activity(GO:0051351) |
| 0.4 | 3.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.4 | 1.8 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.4 | 4.9 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.3 | 1.4 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.3 | 2.8 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.3 | 1.4 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.3 | 1.7 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.3 | 1.3 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.3 | 0.3 | GO:0051095 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.3 | 0.7 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.3 | 1.0 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.3 | 1.2 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.3 | 3.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.3 | 2.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.3 | 1.5 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.3 | 1.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.3 | 0.9 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.3 | 2.4 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.3 | 1.2 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.3 | 1.5 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.3 | 0.9 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.3 | 1.1 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.3 | 1.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.3 | 1.7 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.3 | 1.9 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.3 | 0.8 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.3 | 1.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.3 | 2.1 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.3 | 0.8 | GO:1901534 | positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.3 | 2.5 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.3 | 1.0 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.2 | 1.2 | GO:0032615 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.2 | 0.7 | GO:0090281 | negative regulation of vascular permeability(GO:0043116) negative regulation of calcium ion import(GO:0090281) |
| 0.2 | 0.9 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.2 | 1.6 | GO:0060148 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.2 | 3.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.2 | 0.7 | GO:0021846 | cell proliferation in forebrain(GO:0021846) centriole elongation(GO:0061511) |
| 0.2 | 1.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 0.6 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 0.6 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.2 | 0.6 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.2 | 1.7 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.2 | 1.7 | GO:1903076 | regulation of protein localization to plasma membrane(GO:1903076) regulation of protein localization to cell periphery(GO:1904375) |
| 0.2 | 2.7 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.2 | 0.8 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.2 | 0.6 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.2 | 1.2 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.2 | 1.6 | GO:1901031 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.2 | 0.6 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.2 | 0.8 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.2 | 6.2 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.2 | 1.4 | GO:0045453 | bone resorption(GO:0045453) |
| 0.2 | 0.8 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.2 | 0.8 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.2 | 0.6 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) |
| 0.2 | 0.6 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.2 | 2.6 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.2 | 0.4 | GO:2000058 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.2 | 1.7 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 1.1 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) regulation of fatty acid oxidation(GO:0046320) |
| 0.2 | 0.5 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.2 | 1.2 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.2 | 0.7 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.2 | 3.3 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.2 | 0.5 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.2 | 1.0 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.2 | 0.7 | GO:0003262 | endocardial progenitor cell migration to the midline involved in heart field formation(GO:0003262) |
| 0.2 | 1.3 | GO:0014009 | glial cell proliferation(GO:0014009) regulation of glial cell proliferation(GO:0060251) |
| 0.2 | 3.1 | GO:0032885 | regulation of polysaccharide biosynthetic process(GO:0032885) |
| 0.2 | 2.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.2 | 0.5 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 1.3 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.2 | 1.8 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.2 | 1.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.2 | 0.5 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 0.6 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 0.5 | GO:0042126 | nitrate metabolic process(GO:0042126) nitrate assimilation(GO:0042128) |
| 0.2 | 0.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 0.8 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 1.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.9 | GO:0045981 | positive regulation of nucleotide metabolic process(GO:0045981) positive regulation of purine nucleotide metabolic process(GO:1900544) |
| 0.1 | 0.4 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 1.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 10.7 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 1.3 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 1.1 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.1 | 0.7 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.1 | GO:1901255 | nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.1 | 0.6 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 1.2 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 0.7 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.4 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.1 | 6.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 1.5 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 2.2 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 4.3 | GO:0035825 | reciprocal DNA recombination(GO:0035825) |
| 0.1 | 1.0 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.1 | 1.0 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 0.4 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.8 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.8 | GO:0044033 | multi-organism metabolic process(GO:0044033) |
| 0.1 | 4.3 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.1 | 0.9 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 0.1 | GO:0051341 | regulation of oxidoreductase activity(GO:0051341) |
| 0.1 | 0.4 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.1 | 0.4 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 2.8 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 0.2 | GO:0048714 | positive regulation of gliogenesis(GO:0014015) positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 0.6 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.7 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 1.3 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 0.7 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.1 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.1 | 3.9 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) positive regulation of proteolysis involved in cellular protein catabolic process(GO:1903052) positive regulation of cellular protein catabolic process(GO:1903364) |
| 0.1 | 0.6 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.1 | 3.0 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 0.9 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 1.8 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.4 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.6 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.3 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.1 | 0.2 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 1.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 0.6 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.1 | 0.7 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.3 | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.1 | 0.7 | GO:0043901 | negative regulation of multi-organism process(GO:0043901) |
| 0.1 | 1.6 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 1.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 0.3 | GO:0090022 | neutrophil extravasation(GO:0072672) regulation of neutrophil chemotaxis(GO:0090022) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.1 | 0.5 | GO:0042985 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.1 | 8.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 1.8 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.1 | 1.3 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.1 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.1 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.1 | 0.5 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.1 | 0.7 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 1.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 1.3 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 1.2 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 1.5 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 1.5 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 1.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.5 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.1 | 0.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 1.8 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.1 | 0.6 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.8 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.1 | 0.3 | GO:0038007 | substrate-dependent cell migration(GO:0006929) netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.5 | GO:0021744 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.1 | 0.5 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.1 | 1.5 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 0.3 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 2.5 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.6 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.1 | 0.5 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.1 | 1.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 4.1 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 0.3 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.8 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.3 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 3.1 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 0.2 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.1 | 0.8 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 2.5 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.1 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 1.0 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.6 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.5 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 0.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.4 | GO:0010469 | regulation of receptor activity(GO:0010469) |
| 0.1 | 0.7 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.3 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 0.1 | 1.7 | GO:0007568 | aging(GO:0007568) |
| 0.1 | 0.3 | GO:2000737 | negative regulation of stem cell differentiation(GO:2000737) |
| 0.1 | 0.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.2 | GO:0045161 | neuronal ion channel clustering(GO:0045161) clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.2 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.1 | 1.5 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.1 | 0.5 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.1 | 3.6 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.1 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.9 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 1.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 1.4 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.2 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 1.5 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.1 | 0.4 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.1 | 0.6 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.3 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) |
| 0.1 | 1.2 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 1.0 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.4 | GO:0033750 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.1 | 1.3 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.4 | GO:0003190 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.1 | 1.0 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.3 | GO:0034035 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.3 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 0.3 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.1 | 0.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 1.7 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.1 | 3.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 0.9 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 1.1 | GO:0010833 | telomere maintenance via telomere lengthening(GO:0010833) |
| 0.1 | 1.1 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.1 | 1.4 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.1 | 0.3 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.8 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.5 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.5 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.4 | GO:0061316 | canonical Wnt signaling pathway involved in heart development(GO:0061316) |
| 0.0 | 1.5 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.7 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.1 | GO:0002448 | mast cell activation involved in immune response(GO:0002279) mast cell mediated immunity(GO:0002448) regulation of myeloid leukocyte mediated immunity(GO:0002886) regulation of mast cell activation(GO:0033003) regulation of mast cell activation involved in immune response(GO:0033006) leukocyte degranulation(GO:0043299) regulation of leukocyte degranulation(GO:0043300) mast cell degranulation(GO:0043303) regulation of mast cell degranulation(GO:0043304) |
| 0.0 | 0.1 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.2 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.3 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.2 | GO:1990379 | lysophospholipid transport(GO:0051977) lipid transport across blood brain barrier(GO:1990379) |
| 0.0 | 0.5 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.6 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 1.3 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.5 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.1 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.6 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.8 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.7 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.2 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.1 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) regulation of cellular amine metabolic process(GO:0033238) |
| 0.0 | 1.5 | GO:0048278 | membrane docking(GO:0022406) vesicle docking(GO:0048278) |
| 0.0 | 0.1 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.0 | 0.2 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 0.5 | GO:0036302 | atrioventricular canal development(GO:0036302) |
| 0.0 | 0.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.0 | 0.8 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.3 | GO:0071450 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.2 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.4 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.6 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.7 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0043650 | dicarboxylic acid biosynthetic process(GO:0043650) |
| 0.0 | 0.1 | GO:0002504 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.5 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.0 | 0.6 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 3.6 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 1.0 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0034661 | rRNA catabolic process(GO:0016075) ncRNA catabolic process(GO:0034661) |
| 0.0 | 0.5 | GO:0050852 | antigen receptor-mediated signaling pathway(GO:0050851) T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.2 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 12.6 | GO:0051603 | proteolysis involved in cellular protein catabolic process(GO:0051603) |
| 0.0 | 0.2 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.0 | 0.1 | GO:0061162 | polarized epithelial cell differentiation(GO:0030859) establishment of apical/basal cell polarity(GO:0035089) establishment of epithelial cell apical/basal polarity(GO:0045198) establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.7 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.2 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.2 | GO:0008210 | estrogen biosynthetic process(GO:0006703) estrogen metabolic process(GO:0008210) |
| 0.0 | 0.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.8 | GO:0009306 | protein secretion(GO:0009306) |
| 0.0 | 0.2 | GO:0060969 | negative regulation of gene silencing(GO:0060969) |
| 0.0 | 0.2 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.2 | GO:0006901 | vesicle coating(GO:0006901) |
| 0.0 | 0.4 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 1.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 0.5 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.4 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 | 0.3 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) |
| 0.0 | 0.5 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.4 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.8 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.3 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.0 | GO:0071548 | response to dexamethasone(GO:0071548) |
| 0.0 | 0.1 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 1.2 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.2 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 1.3 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0000423 | macromitophagy(GO:0000423) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 1.7 | GO:0006310 | DNA recombination(GO:0006310) |
| 0.0 | 0.7 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.1 | GO:2000765 | regulation of cytoplasmic translation(GO:2000765) |
| 0.0 | 0.1 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.3 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 1.0 | GO:0007346 | regulation of mitotic cell cycle(GO:0007346) |
| 0.0 | 0.3 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 1.6 | GO:0007276 | gamete generation(GO:0007276) |
| 0.0 | 0.0 | GO:0030817 | regulation of cyclic nucleotide metabolic process(GO:0030799) regulation of cyclic nucleotide biosynthetic process(GO:0030802) regulation of nucleotide biosynthetic process(GO:0030808) regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of cyclase activity(GO:0031279) regulation of adenylate cyclase activity(GO:0045761) regulation of lyase activity(GO:0051339) regulation of purine nucleotide biosynthetic process(GO:1900371) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.4 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.1 | GO:0010677 | negative regulation of cellular carbohydrate metabolic process(GO:0010677) negative regulation of gluconeogenesis(GO:0045721) negative regulation of carbohydrate metabolic process(GO:0045912) |
| 0.0 | 0.4 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.5 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.1 | GO:0009204 | deoxyribonucleoside triphosphate catabolic process(GO:0009204) |
| 0.0 | 0.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.0 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.0 | 0.3 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 1.4 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.1 | GO:0006497 | protein lipidation(GO:0006497) lipoprotein biosynthetic process(GO:0042158) |
| 0.0 | 0.1 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.1 | GO:0009746 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.0 | 0.6 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.3 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.9 | GO:0042382 | paraspeckles(GO:0042382) |
| 1.0 | 2.9 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.9 | 3.5 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.5 | 2.7 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.5 | 2.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.5 | 1.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.5 | 5.6 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.5 | 1.9 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.5 | 1.4 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.4 | 3.0 | GO:0071546 | pi-body(GO:0071546) |
| 0.4 | 1.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.4 | 1.7 | GO:0098536 | deuterosome(GO:0098536) |
| 0.4 | 1.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.4 | 1.2 | GO:0097361 | CIA complex(GO:0097361) |
| 0.4 | 1.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.4 | 1.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.4 | 1.8 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.4 | 2.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.3 | 3.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.3 | 4.6 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.3 | 2.9 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.3 | 1.0 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.3 | 1.5 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.3 | 3.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.3 | 3.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.3 | 1.7 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.3 | 1.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.3 | 1.1 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.3 | 3.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.3 | 0.8 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.3 | 2.5 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.3 | 1.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 0.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 2.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 1.2 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 1.6 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 2.7 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 2.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 2.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 1.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 0.6 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.2 | 1.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 0.6 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.2 | 0.5 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.2 | 3.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 2.0 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 2.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 0.6 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.2 | 2.1 | GO:0015030 | Cajal body(GO:0015030) |
| 0.2 | 0.9 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.2 | 0.8 | GO:0000811 | GINS complex(GO:0000811) |
| 0.1 | 2.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 5.8 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.3 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 0.7 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 1.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 1.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 3.0 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 2.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.7 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.1 | 1.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 1.7 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 0.8 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 1.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 3.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 0.4 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.1 | 0.5 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.5 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.1 | 0.5 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 4.1 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.1 | 0.8 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 3.1 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.1 | 2.2 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 0.8 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.1 | 0.3 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.5 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 0.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 0.3 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.1 | 1.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 2.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.4 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 2.4 | GO:0071010 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.1 | 2.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 2.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.7 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.7 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 3.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.5 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.1 | 1.2 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 0.5 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 1.0 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 0.7 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 5.5 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.1 | 0.7 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.4 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 7.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 3.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.2 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 3.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 0.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.8 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 0.2 | GO:0097196 | Shu complex(GO:0097196) |
| 0.1 | 1.2 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.1 | 0.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.6 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.5 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.1 | 1.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 3.0 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.1 | 0.4 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 1.7 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 1.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.2 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 0.3 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 10.9 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 3.3 | GO:0035770 | ribonucleoprotein granule(GO:0035770) cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 0.4 | GO:0097346 | INO80-type complex(GO:0097346) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 3.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.3 | GO:0036452 | ESCRT complex(GO:0036452) |
| 0.0 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.3 | GO:0072379 | ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.3 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 1.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 2.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.1 | GO:0034991 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 1.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 3.2 | GO:0000786 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.0 | 0.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.8 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 2.2 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 2.6 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.4 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.0 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.9 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.5 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.5 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.0 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 10.3 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.2 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 1.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.0 | GO:0005774 | vacuolar membrane(GO:0005774) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.1 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 3.9 | GO:0005694 | chromosome(GO:0005694) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0047611 | acetylspermidine deacetylase activity(GO:0047611) |
| 1.0 | 4.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 1.0 | 3.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 1.0 | 3.0 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 1.0 | 6.9 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.9 | 2.8 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.8 | 2.5 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.8 | 3.0 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.7 | 2.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.7 | 4.4 | GO:0090624 | endoribonuclease activity, cleaving miRNA-paired mRNA(GO:0090624) |
| 0.7 | 2.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.7 | 2.7 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.6 | 1.8 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.6 | 2.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.6 | 1.7 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.5 | 4.9 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.5 | 2.1 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.5 | 2.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.5 | 1.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.5 | 9.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.5 | 6.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.5 | 5.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.5 | 1.9 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.5 | 1.4 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.5 | 2.3 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.4 | 3.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.4 | 1.8 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.4 | 1.7 | GO:0019705 | protein-cysteine S-myristoyltransferase activity(GO:0019705) |
| 0.4 | 4.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.4 | 1.7 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.4 | 5.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.4 | 2.3 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.4 | 3.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.4 | 1.9 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.4 | 1.1 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.4 | 1.1 | GO:0071424 | rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) |
| 0.4 | 1.4 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.4 | 1.1 | GO:0031420 | pyruvate kinase activity(GO:0004743) potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.3 | 1.4 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.3 | 5.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.3 | 11.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.3 | 1.9 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.3 | 1.0 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.3 | 2.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 1.2 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.3 | 1.5 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.3 | 1.2 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.3 | 0.6 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.3 | 3.3 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.3 | 1.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.3 | 1.2 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.3 | 0.8 | GO:0004948 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) calcitonin receptor activity(GO:0004948) calcitonin family receptor activity(GO:0097642) |
| 0.3 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.3 | 1.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 2.1 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.2 | 1.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.2 | 1.4 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.2 | 1.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 0.7 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.2 | 1.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 1.9 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 4.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 3.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 1.0 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 1.6 | GO:0070728 | leucine binding(GO:0070728) |
| 0.2 | 3.0 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 0.4 | GO:0001006 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 3 promoter sequence-specific DNA binding(GO:0001006) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.2 | 3.4 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.2 | 2.6 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.2 | 1.7 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.2 | 1.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 0.7 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 3.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 1.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 1.4 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 2.9 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 0.5 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 0.5 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.2 | 1.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.2 | 0.6 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 0.5 | GO:0008942 | nitrite reductase [NAD(P)H] activity(GO:0008942) oxidoreductase activity, acting on other nitrogenous compounds as donors, with NAD or NADP as acceptor(GO:0046857) |
| 0.2 | 0.9 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.2 | 0.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.9 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 5.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.4 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.1 | 0.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 3.2 | GO:0016896 | exoribonuclease activity(GO:0004532) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.1 | 1.0 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.7 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 0.5 | GO:0015198 | oligopeptide transporter activity(GO:0015198) oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.9 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.8 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.9 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 0.8 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 2.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.4 | GO:0002094 | polyprenyltransferase activity(GO:0002094) dehydrodolichyl diphosphate synthase activity(GO:0045547) |
| 0.1 | 0.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.8 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 1.0 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 0.2 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.1 | 1.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 1.3 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 0.5 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 1.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 2.1 | GO:0004407 | histone deacetylase activity(GO:0004407) NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.1 | 1.0 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 2.4 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.1 | 2.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 3.7 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 0.7 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.1 | 1.9 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 2.0 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.1 | 1.1 | GO:0097200 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 1.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 1.2 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.4 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 2.1 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 3.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.0 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.4 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.1 | 0.5 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.1 | 0.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.7 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 1.3 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 6.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 0.6 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 1.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 0.6 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 0.4 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 3.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 1.5 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 1.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 3.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 1.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.8 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.7 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 1.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 6.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 1.1 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.1 | 1.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 2.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 27.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 0.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 1.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.7 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.1 | 1.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.9 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.4 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 0.5 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.3 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 1.1 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.6 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.5 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 1.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.3 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 2.6 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 1.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.5 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 1.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 0.5 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 0.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.2 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.4 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.2 | GO:0051978 | lysophospholipid transporter activity(GO:0051978) |
| 0.0 | 2.2 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.8 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 3.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.8 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 1.0 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 2.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 3.1 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 1.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.4 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 3.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.6 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.2 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.1 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.0 | 7.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 1.0 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 3.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.1 | GO:0043878 | aminobutyraldehyde dehydrogenase activity(GO:0019145) glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 1.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 2.4 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.4 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.3 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 1.1 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.2 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.1 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.5 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) RNA polymerase binding(GO:0070063) |
| 0.0 | 0.2 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 2.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.3 | GO:0019787 | ubiquitin-like protein transferase activity(GO:0019787) |
| 0.0 | 0.3 | GO:0015645 | fatty acid ligase activity(GO:0015645) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.0 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.3 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0043394 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.5 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.0 | GO:0003874 | 6-pyruvoyltetrahydropterin synthase activity(GO:0003874) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.5 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.4 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.6 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.6 | 3.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.6 | 1.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.4 | 0.4 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.3 | 2.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.2 | 8.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.2 | 1.9 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 3.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 0.5 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 4.3 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 3.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 4.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 0.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 3.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 0.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.5 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 3.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 1.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 4.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 0.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 5.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.1 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 1.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.1 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 3.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 2.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 0.9 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 0.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.9 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.2 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.0 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 0.9 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.6 | 4.5 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.5 | 2.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.5 | 5.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.4 | 2.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.3 | 2.1 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.3 | 2.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 9.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.3 | 2.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.2 | 4.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.2 | 3.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 2.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 3.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.2 | 3.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 0.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 1.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.2 | 6.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 0.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 1.9 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.2 | 2.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.2 | 1.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 2.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 6.9 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.1 | 2.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 1.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 1.4 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 0.4 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.1 | 1.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 3.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 1.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 1.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.4 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 0.5 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 1.0 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.2 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.1 | 1.8 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.1 | 0.5 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 1.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 4.7 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.1 | 0.6 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.1 | 0.8 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 0.8 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.1 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 0.7 | REACTOME METABOLISM OF RNA | Genes involved in Metabolism of RNA |
| 0.1 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 0.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 2.1 | REACTOME RNA POL III TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.1 | 0.9 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 0.6 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 0.3 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 10.8 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 0.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 0.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 2.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 2.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.5 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 1.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.8 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.6 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.5 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.6 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME SHC MEDIATED CASCADE | Genes involved in SHC-mediated cascade |
| 0.0 | 0.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME MEMBRANE TRAFFICKING | Genes involved in Membrane Trafficking |
| 0.0 | 0.2 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.1 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |