Project
DANIO-CODE
Navigation
Downloads
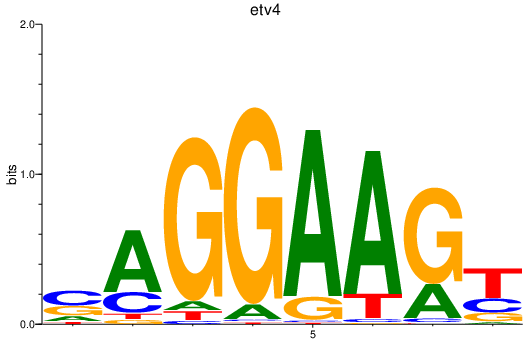
Results for etv4
Z-value: 0.85
Transcription factors associated with etv4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
etv4
|
ENSDARG00000018303 | ETS variant transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| etv4 | dr10_dc_chr12_+_27444732_27444735 | -0.78 | 3.6e-04 | Click! |
Activity profile of etv4 motif
Sorted Z-values of etv4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of etv4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_11912981 | 0.84 |
ENSDART00000158244
|
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr20_+_7596461 | 0.67 |
ENSDART00000127975
ENSDART00000132481 ENSDART00000144551 |
bloc1s2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr13_+_11913290 | 0.64 |
ENSDART00000079398
|
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr7_+_19230972 | 0.63 |
ENSDART00000077868
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr7_-_26767182 | 0.62 |
ENSDART00000052731
|
nucb2a
|
nucleobindin 2a |
| chr23_+_10211543 | 0.61 |
ENSDART00000048073
|
zgc:171775
|
zgc:171775 |
| chr14_+_23412305 | 0.60 |
ENSDART00000136909
|
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr7_-_48465949 | 0.57 |
ENSDART00000038273
|
traf6
|
TNF receptor-associated factor 6 |
| chr13_+_30939809 | 0.56 |
ENSDART00000110560
|
si:ch211-223a10.1
|
si:ch211-223a10.1 |
| chr6_-_54172509 | 0.56 |
ENSDART00000149468
|
rps10
|
ribosomal protein S10 |
| KN150680v1_+_10854 | 0.55 |
|
|
|
| chr3_+_16115708 | 0.55 |
ENSDART00000122519
|
st8sia6
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
| chr21_-_44013995 | 0.55 |
ENSDART00000028960
|
ndufa2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 |
| chr17_-_43041127 | 0.55 |
ENSDART00000132754
|
npc2
|
Niemann-Pick disease, type C2 |
| chr15_-_29393252 | 0.55 |
ENSDART00000114492
|
si:dkey-52l18.4
|
si:dkey-52l18.4 |
| chr17_+_3842688 | 0.54 |
ENSDART00000170822
|
tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr8_-_7049946 | 0.54 |
ENSDART00000045669
ENSDART00000162950 |
KRT73
|
keratin 73 |
| chr14_+_49921861 | 0.53 |
ENSDART00000173240
|
zgc:154054
|
zgc:154054 |
| chr15_-_4537178 | 0.53 |
ENSDART00000155619
ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr9_-_711269 | 0.53 |
ENSDART00000144625
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr15_+_36587375 | 0.52 |
ENSDART00000154552
|
si:dkey-262k9.2
|
si:dkey-262k9.2 |
| chr14_+_38538151 | 0.52 |
ENSDART00000043317
|
timm8a
|
translocase of inner mitochondrial membrane 8 homolog A (yeast) |
| chr9_-_21427331 | 0.52 |
ENSDART00000102143
|
cox17
|
COX17 cytochrome c oxidase copper chaperone |
| chr1_-_55108003 | 0.50 |
ENSDART00000142069
ENSDART00000043933 |
ndufb7
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7 |
| chr14_+_24637864 | 0.50 |
ENSDART00000170871
|
ppargc1b
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr12_+_27370834 | 0.48 |
ENSDART00000105661
|
meox1
|
mesenchyme homeobox 1 |
| chr20_+_31366832 | 0.48 |
ENSDART00000133353
|
apobb.1
|
apolipoprotein Bb, tandem duplicate 1 |
| chr1_-_48790379 | 0.47 |
ENSDART00000150842
|
USMG5
|
up-regulated during skeletal muscle growth 5 homolog (mouse) |
| chr19_-_36146250 | 0.46 |
ENSDART00000066712
|
trappc3
|
trafficking protein particle complex 3 |
| chr10_-_42302932 | 0.45 |
ENSDART00000076693
ENSDART00000073631 |
stambpa
|
STAM binding protein a |
| chr15_+_30430093 | 0.45 |
ENSDART00000112784
|
lyrm9
|
LYR motif containing 9 |
| chr20_+_54525614 | 0.45 |
ENSDART00000160409
|
arf6a
|
ADP-ribosylation factor 6a |
| chr7_+_67244332 | 0.44 |
ENSDART00000170322
|
rpl13
|
ribosomal protein L13 |
| chr14_-_38538068 | 0.44 |
ENSDART00000173300
|
uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr11_-_25803101 | 0.43 |
ENSDART00000088888
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr19_-_18689437 | 0.43 |
ENSDART00000016135
|
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr20_+_20600211 | 0.43 |
ENSDART00000036124
|
six1b
|
SIX homeobox 1b |
| chr19_-_325337 | 0.43 |
ENSDART00000147635
|
gpd1c
|
glycerol-3-phosphate dehydrogenase 1c |
| chr3_-_32770358 | 0.43 |
ENSDART00000145443
|
kat7a
|
K(lysine) acetyltransferase 7a |
| chr6_+_38882803 | 0.43 |
ENSDART00000132798
|
bin2b
|
bridging integrator 2b |
| chr19_-_11290766 | 0.42 |
ENSDART00000044426
|
si:dkey-240h12.4
|
si:dkey-240h12.4 |
| chr10_-_13814731 | 0.42 |
ENSDART00000145103
|
cntfr
|
ciliary neurotrophic factor receptor |
| chr24_-_13205000 | 0.42 |
ENSDART00000134482
|
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr15_-_9655500 | 0.42 |
ENSDART00000171214
ENSDART00000171911 |
nars2
|
asparaginyl-tRNA synthetase 2, mitochondrial (putative) |
| chr9_-_24013113 | 0.42 |
ENSDART00000059209
|
wdfy2
|
WD repeat and FYVE domain containing 2 |
| chr4_-_978592 | 0.41 |
ENSDART00000160902
|
naga
|
N-acetylgalactosaminidase, alpha |
| chr9_-_1964814 | 0.41 |
ENSDART00000082354
|
hoxd9a
|
homeobox D9a |
| chr3_+_31542311 | 0.41 |
|
|
|
| chr15_+_20345529 | 0.40 |
ENSDART00000155065
|
plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr22_-_120677 | 0.40 |
|
|
|
| chr3_+_27684309 | 0.39 |
|
|
|
| chr15_-_29393320 | 0.39 |
ENSDART00000114492
|
si:dkey-52l18.4
|
si:dkey-52l18.4 |
| chr18_-_1076316 | 0.39 |
ENSDART00000134144
|
ENSDARG00000019081
|
ENSDARG00000019081 |
| chr25_+_14411153 | 0.39 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr17_+_15205550 | 0.38 |
ENSDART00000058351
|
gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr25_+_7660590 | 0.38 |
ENSDART00000155016
|
dgkzb
|
diacylglycerol kinase, zeta b |
| chr5_+_23741791 | 0.38 |
ENSDART00000049003
|
atp6v1aa
|
ATPase, H+ transporting, lysosomal, V1 subunit Aa |
| chr7_-_29906486 | 0.38 |
ENSDART00000046689
|
tmed3
|
transmembrane p24 trafficking protein 3 |
| chr6_-_12015393 | 0.37 |
|
|
|
| chr4_+_3344998 | 0.37 |
ENSDART00000075320
|
nampta
|
nicotinamide phosphoribosyltransferase a |
| chr23_+_36007936 | 0.37 |
ENSDART00000128533
|
hoxc3a
|
homeo box C3a |
| chr18_-_5392973 | 0.37 |
ENSDART00000082496
|
dtwd1
|
DTW domain containing 1 |
| chr15_-_11798226 | 0.37 |
|
|
|
| chr16_-_36991800 | 0.37 |
ENSDART00000171351
|
aldh5a1
|
aldehyde dehydrogenase 5 family, member A1 (succinate-semialdehyde dehydrogenase) |
| chr17_-_7635061 | 0.37 |
ENSDART00000064655
|
zbtb2a
|
zinc finger and BTB domain containing 2a |
| chr9_+_13149212 | 0.37 |
ENSDART00000141005
|
fam117bb
|
family with sequence similarity 117, member Bb |
| chr3_+_24510920 | 0.37 |
ENSDART00000148414
ENSDART00000055590 |
zgc:113411
|
zgc:113411 |
| chr16_+_48729441 | 0.36 |
ENSDART00000154683
|
CU694532.1
|
ENSDARG00000097878 |
| chr6_-_19556328 | 0.36 |
ENSDART00000074256
|
ppp1r12c
|
protein phosphatase 1, regulatory subunit 12C |
| chr5_-_48377307 | 0.36 |
ENSDART00000097452
|
si:dkey-172m14.1
|
si:dkey-172m14.1 |
| chr9_-_7560915 | 0.35 |
ENSDART00000081553
|
desma
|
desmin a |
| chr8_+_30998992 | 0.35 |
ENSDART00000129882
|
ptgesl
|
prostaglandin E synthase 2-like |
| chr5_-_1325831 | 0.35 |
|
|
|
| chr9_+_56881036 | 0.35 |
|
|
|
| chr25_-_3633631 | 0.35 |
ENSDART00000159335
ENSDART00000088077 |
zgc:158398
|
zgc:158398 |
| chr19_-_2399429 | 0.35 |
ENSDART00000043595
|
twist1a
|
twist family bHLH transcription factor 1a |
| chr17_-_43725003 | 0.35 |
|
|
|
| chr3_+_52974018 | 0.34 |
ENSDART00000156490
|
pet100
|
PET100 homolog |
| chr12_+_22448972 | 0.34 |
|
|
|
| chr6_+_36861678 | 0.34 |
ENSDART00000104160
|
ENSDARG00000070746
|
ENSDARG00000070746 |
| chr14_-_20940726 | 0.34 |
ENSDART00000129743
|
si:ch211-175m2.5
|
si:ch211-175m2.5 |
| chr9_-_31714290 | 0.34 |
ENSDART00000127214
|
tmtc4
|
transmembrane and tetratricopeptide repeat containing 4 |
| chr21_+_16987306 | 0.33 |
ENSDART00000080628
|
arpc3
|
actin related protein 2/3 complex, subunit 3 |
| chr6_-_3837294 | 0.33 |
ENSDART00000059212
|
unc50
|
unc-50 homolog (C. elegans) |
| chr14_-_23953804 | 0.33 |
ENSDART00000114169
|
bnip1a
|
BCL2/adenovirus E1B interacting protein 1a |
| chr17_-_49895398 | 0.33 |
ENSDART00000155446
|
cox7a2a
|
cytochrome c oxidase subunit VIIa polypeptide 2a (liver) |
| chr11_+_8119829 | 0.33 |
ENSDART00000011183
|
prkacba
|
protein kinase, cAMP-dependent, catalytic, beta a |
| chr10_+_16589111 | 0.33 |
ENSDART00000084278
|
isoc1
|
isochorismatase domain containing 1 |
| chr20_-_42821812 | 0.32 |
ENSDART00000138957
|
gpr31
|
G protein-coupled receptor 31 |
| chr20_-_3074186 | 0.32 |
ENSDART00000046641
|
map3k5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr18_-_18535519 | 0.32 |
ENSDART00000157834
|
lonp2
|
lon peptidase 2, peroxisomal |
| chr13_+_3533517 | 0.32 |
ENSDART00000018737
|
qkib
|
QKI, KH domain containing, RNA binding b |
| chr8_-_49356479 | 0.31 |
ENSDART00000053203
|
plp2
|
proteolipid protein 2 |
| chr24_-_9819862 | 0.31 |
ENSDART00000092975
|
vps41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr1_+_50763649 | 0.31 |
ENSDART00000074294
|
actr2a
|
ARP2 actin related protein 2a homolog |
| chr10_+_13251463 | 0.31 |
ENSDART00000000887
|
rassf6
|
Ras association (RalGDS/AF-6) domain family 6 |
| chr8_+_54171992 | 0.31 |
ENSDART00000122692
|
CABZ01112317.1
|
ENSDARG00000086057 |
| chr3_-_23466232 | 0.30 |
ENSDART00000156897
|
ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr1_+_38099962 | 0.30 |
ENSDART00000166864
|
spcs3
|
signal peptidase complex subunit 3 |
| chr10_+_24721186 | 0.30 |
ENSDART00000144710
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr10_+_4987494 | 0.30 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr24_+_12839000 | 0.30 |
ENSDART00000145214
|
tceb1a
|
transcription elongation factor B (SIII), polypeptide 1a |
| chr22_-_18724510 | 0.30 |
ENSDART00000167466
|
midn
|
midnolin |
| chr11_+_5929544 | 0.30 |
ENSDART00000104364
|
rps15
|
ribosomal protein S15 |
| chr11_+_37639045 | 0.29 |
ENSDART00000111157
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr17_-_30958965 | 0.29 |
ENSDART00000051697
|
evla
|
Enah/Vasp-like a |
| chr10_-_36849108 | 0.29 |
|
|
|
| chr3_+_24990797 | 0.29 |
ENSDART00000025724
|
ENSDARG00000009313
|
ENSDARG00000009313 |
| chr19_+_17470263 | 0.29 |
ENSDART00000160478
ENSDART00000158935 |
clic1
|
chloride intracellular channel 1 |
| chr5_+_7691621 | 0.29 |
ENSDART00000161261
|
lmbrd2a
|
LMBR1 domain containing 2a |
| chr23_+_22729939 | 0.29 |
ENSDART00000009337
|
eno1a
|
enolase 1a, (alpha) |
| chr23_+_19749426 | 0.29 |
|
|
|
| chr16_+_5470544 | 0.29 |
|
|
|
| chr20_+_13279404 | 0.28 |
ENSDART00000025644
|
ppp2r5a
|
protein phosphatase 2, regulatory subunit B', alpha isoform |
| chr10_+_4987461 | 0.28 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr10_-_27261937 | 0.28 |
|
|
|
| chr2_-_31728072 | 0.28 |
ENSDART00000113498
|
lrrcc1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr20_+_16843502 | 0.28 |
ENSDART00000050308
|
calm1b
|
calmodulin 1b |
| chr7_-_53828910 | 0.28 |
ENSDART00000165499
|
rps17
|
ribosomal protein S17 |
| chr25_+_22489716 | 0.28 |
ENSDART00000127831
|
stra6
|
stimulated by retinoic acid 6 |
| KN149842v1_+_13298 | 0.27 |
ENSDART00000166541
|
CABZ01101812.1
|
ENSDARG00000098297 |
| chr19_+_19620893 | 0.27 |
ENSDART00000161570
|
mrpl3
|
mitochondrial ribosomal protein L3 |
| chr14_-_47073113 | 0.27 |
|
|
|
| chr15_-_31296524 | 0.27 |
ENSDART00000008854
|
wsb1
|
WD repeat and SOCS box containing 1 |
| chr3_+_25933164 | 0.27 |
ENSDART00000143697
|
si:dkeyp-69e1.8
|
si:dkeyp-69e1.8 |
| chr22_+_31089813 | 0.27 |
ENSDART00000060005
|
rpl32
|
ribosomal protein L32 |
| chr6_-_33940469 | 0.27 |
ENSDART00000137268
|
akr1a1b
|
aldo-keto reductase family 1, member A1b (aldehyde reductase) |
| chr8_+_15201828 | 0.27 |
ENSDART00000132216
|
paox1
|
polyamine oxidase (exo-N4-amino) 1 |
| chr11_-_41750987 | 0.27 |
ENSDART00000166999
|
zgc:175088
|
zgc:175088 |
| KN149909v1_+_1319 | 0.27 |
|
|
|
| chr3_-_39313029 | 0.27 |
ENSDART00000156123
|
CU464118.1
|
ENSDARG00000097747 |
| chr8_-_49356695 | 0.27 |
ENSDART00000053203
|
plp2
|
proteolipid protein 2 |
| chr14_+_6639864 | 0.26 |
ENSDART00000061001
|
gnb2l1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr2_-_32591634 | 0.26 |
ENSDART00000056642
|
tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr24_+_31340115 | 0.26 |
ENSDART00000165993
|
f3a
|
coagulation factor IIIa |
| chr23_+_34030832 | 0.26 |
ENSDART00000172069
|
snrpg
|
small nuclear ribonucleoprotein polypeptide G |
| chr24_-_40968409 | 0.26 |
ENSDART00000169315
ENSDART00000171543 |
smyhc1
|
slow myosin heavy chain 1 |
| chr1_+_9633924 | 0.26 |
ENSDART00000029774
|
tmem55bb
|
transmembrane protein 55Bb |
| chr10_+_22759607 | 0.25 |
|
|
|
| chr15_-_5827067 | 0.25 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr6_-_19556121 | 0.25 |
ENSDART00000136019
|
ppp1r12c
|
protein phosphatase 1, regulatory subunit 12C |
| chr17_+_10086279 | 0.25 |
ENSDART00000170420
|
clec14a
|
C-type lectin domain containing 14A |
| chr1_-_10122492 | 0.25 |
ENSDART00000139749
|
ENSDARG00000087107
|
ENSDARG00000087107 |
| chr20_+_52740555 | 0.25 |
ENSDART00000110777
|
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr13_+_23065500 | 0.25 |
ENSDART00000158370
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr7_+_23752492 | 0.25 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr10_-_13281047 | 0.25 |
ENSDART00000001253
|
si:busm1-57f23.1
|
si:busm1-57f23.1 |
| chr1_-_44227583 | 0.25 |
ENSDART00000134635
|
si:dkey-9i23.15
|
si:dkey-9i23.15 |
| chr7_+_28844711 | 0.25 |
ENSDART00000052345
|
tradd
|
tnfrsf1a-associated via death domain |
| chr22_-_23641813 | 0.25 |
ENSDART00000159622
|
cfh
|
complement factor H |
| chr14_-_47210912 | 0.25 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr2_-_10020770 | 0.25 |
ENSDART00000046587
|
ap2m1a
|
adaptor-related protein complex 2, mu 1 subunit, a |
| chr22_+_26616977 | 0.25 |
ENSDART00000171972
ENSDART00000083351 |
ENSDARG00000034211
|
ENSDARG00000034211 |
| chr11_+_4934132 | 0.24 |
ENSDART00000178630
|
ptprga
|
protein tyrosine phosphatase, receptor type, g a |
| chr23_-_28367816 | 0.24 |
ENSDART00000003548
|
znf385a
|
zinc finger protein 385A |
| chr16_+_13928376 | 0.24 |
ENSDART00000163251
|
flcn
|
folliculin |
| chr20_+_26408687 | 0.24 |
|
|
|
| chr18_+_3482740 | 0.24 |
ENSDART00000158763
|
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr6_+_36862078 | 0.24 |
ENSDART00000104160
|
ENSDARG00000070746
|
ENSDARG00000070746 |
| chr2_+_22839514 | 0.24 |
ENSDART00000167915
|
lrrc8da
|
leucine rich repeat containing 8 family, member Da |
| chr25_+_26962541 | 0.23 |
ENSDART00000115139
|
pot1
|
protection of telomeres 1 homolog |
| chr24_-_6048914 | 0.23 |
ENSDART00000146830
ENSDART00000021981 |
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr3_-_34623996 | 0.23 |
ENSDART00000000160
|
thraa
|
thyroid hormone receptor alpha a |
| chr21_+_20347283 | 0.23 |
ENSDART00000026430
|
hspb11
|
heat shock protein, alpha-crystallin-related, b11 |
| chr3_+_13043951 | 0.23 |
|
|
|
| chr6_-_40715613 | 0.23 |
ENSDART00000153702
|
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr6_-_49064365 | 0.23 |
ENSDART00000156124
|
si:ch211-105j21.9
|
si:ch211-105j21.9 |
| chr6_-_42420644 | 0.23 |
ENSDART00000002501
|
ip6k2a
|
inositol hexakisphosphate kinase 2a |
| chr20_-_47680305 | 0.23 |
ENSDART00000023058
|
efhc1
|
EF-hand domain (C-terminal) containing 1 |
| chr9_+_9524688 | 0.23 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr25_-_35159840 | 0.23 |
ENSDART00000148718
ENSDART00000164189 ENSDART00000027174 |
lrrk2
|
leucine-rich repeat kinase 2 |
| chr12_+_41016238 | 0.23 |
ENSDART00000170976
|
kif5bb
|
kinesin family member 5B, b |
| chr21_-_25258473 | 0.23 |
ENSDART00000101153
ENSDART00000147860 ENSDART00000087910 |
st14b
|
suppression of tumorigenicity 14 (colon carcinoma) b |
| chr6_+_11771219 | 0.23 |
ENSDART00000128024
ENSDART00000121431 |
wdsub1
|
WD repeat, sterile alpha motif and U-box domain containing 1 |
| chr19_+_20405107 | 0.23 |
ENSDART00000151066
|
osbpl3a
|
oxysterol binding protein-like 3a |
| chr13_+_3533318 | 0.22 |
ENSDART00000018737
|
qkib
|
QKI, KH domain containing, RNA binding b |
| chr16_+_21128596 | 0.22 |
ENSDART00000052660
|
skap2
|
src kinase associated phosphoprotein 2 |
| chr19_+_17470304 | 0.22 |
ENSDART00000160478
|
clic1
|
chloride intracellular channel 1 |
| chr21_-_40694583 | 0.22 |
ENSDART00000045124
|
pomp
|
proteasome maturation protein |
| chr20_+_15119519 | 0.22 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr5_-_41672394 | 0.22 |
ENSDART00000164363
|
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr6_-_12224562 | 0.22 |
ENSDART00000090266
ENSDART00000144028 |
gpd2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr10_-_7712439 | 0.22 |
ENSDART00000159330
|
pcyox1
|
prenylcysteine oxidase 1 |
| chr9_-_56644466 | 0.22 |
ENSDART00000149851
|
rpl31
|
ribosomal protein L31 |
| chr2_+_30266427 | 0.22 |
ENSDART00000135171
|
tmem70
|
transmembrane protein 70 |
| chr6_-_33886005 | 0.22 |
ENSDART00000115409
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr8_+_19457907 | 0.22 |
ENSDART00000011258
|
npl
|
N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase) |
| KN150503v1_-_598 | 0.22 |
ENSDART00000174608
|
CABZ01110189.1
|
ENSDARG00000108333 |
| chr19_+_43763483 | 0.22 |
ENSDART00000136695
|
yrk
|
Yes-related kinase |
| chr6_-_33940653 | 0.22 |
ENSDART00000141822
|
akr1a1b
|
aldo-keto reductase family 1, member A1b (aldehyde reductase) |
| chr5_-_60904215 | 0.22 |
ENSDART00000050912
|
pex12
|
peroxisomal biogenesis factor 12 |
| chr11_-_33925792 | 0.22 |
ENSDART00000172978
|
atp13a3
|
ATPase type 13A3 |
| chr20_-_36897042 | 0.22 |
ENSDART00000015190
|
ptrhd1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr16_+_53221957 | 0.22 |
|
|
|
| chr19_-_15288270 | 0.22 |
ENSDART00000151451
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr24_+_21828704 | 0.22 |
ENSDART00000042495
|
sat1b
|
spermidine/spermine N1-acetyltransferase 1b |
| chr17_+_38348229 | 0.22 |
ENSDART00000154346
|
mbip
|
MAP3K12 binding inhibitory protein 1 |
| chr16_+_29574449 | 0.22 |
ENSDART00000148450
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr1_-_44227620 | 0.21 |
ENSDART00000134635
|
si:dkey-9i23.15
|
si:dkey-9i23.15 |
| chr14_+_223907 | 0.21 |
ENSDART00000164411
|
zbtb49
|
zinc finger and BTB domain containing 49 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.2 | 0.6 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 0.1 | 0.4 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 0.6 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 0.6 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.1 | 0.7 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.1 | 0.3 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 0.4 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.5 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.7 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.1 | 0.4 | GO:2001032 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 0.4 | GO:0032615 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.1 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.6 | GO:0032099 | regulation of response to food(GO:0032095) negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.1 | 0.4 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.1 | 0.4 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.5 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.1 | 0.2 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.1 | 0.5 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.6 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 0.2 | GO:0030329 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.1 | 0.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.5 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.1 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.6 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.4 | GO:0060148 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.1 | 0.4 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.4 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.1 | 0.2 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.1 | 0.4 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.1 | 0.4 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.1 | 0.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.2 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.7 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 0.3 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.5 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 1.7 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.4 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) amino sugar biosynthetic process(GO:0046349) |
| 0.0 | 0.1 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.7 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.7 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.7 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:0097037 | heme export(GO:0097037) |
| 0.0 | 0.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.2 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.0 | 0.1 | GO:0032024 | positive regulation of peptide secretion(GO:0002793) positive regulation of insulin secretion(GO:0032024) positive regulation of hormone secretion(GO:0046887) positive regulation of peptide hormone secretion(GO:0090277) |
| 0.0 | 0.3 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.5 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.2 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.1 | GO:0061687 | xenobiotic transport(GO:0042908) detoxification of inorganic compound(GO:0061687) |
| 0.0 | 0.1 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.0 | 0.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.3 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.1 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.0 | 0.1 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.0 | 0.6 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.3 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.1 | GO:0030804 | positive regulation of cyclic nucleotide metabolic process(GO:0030801) positive regulation of cyclic nucleotide biosynthetic process(GO:0030804) positive regulation of nucleotide biosynthetic process(GO:0030810) positive regulation of cyclase activity(GO:0031281) positive regulation of lyase activity(GO:0051349) positive regulation of purine nucleotide biosynthetic process(GO:1900373) |
| 0.0 | 0.1 | GO:0003400 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.0 | 0.3 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.4 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.0 | 0.5 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 0.1 | GO:0043096 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) |
| 0.0 | 0.2 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0051126 | negative regulation of actin nucleation(GO:0051126) |
| 0.0 | 0.3 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) |
| 0.0 | 0.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.1 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.3 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.3 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.1 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.3 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.1 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.4 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.6 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.4 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.3 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.2 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.5 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0036372 | opsin transport(GO:0036372) |
| 0.0 | 0.2 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.3 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.1 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.3 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.0 | GO:0090281 | negative regulation of vascular permeability(GO:0043116) negative regulation of calcium ion import(GO:0090281) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0072599 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.2 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.6 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 0.3 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.0 | 0.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.1 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.0 | GO:0000493 | box H/ACA snoRNP assembly(GO:0000493) |
| 0.0 | 0.2 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.0 | 0.2 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.1 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.1 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.0 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0098594 | mucin granule(GO:0098594) |
| 0.1 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.5 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.7 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.3 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 0.5 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.4 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 0.2 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.6 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 1.9 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 1.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.3 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.9 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.6 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.6 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.5 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.2 | GO:0032587 | ruffle membrane(GO:0032587) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.2 | 0.6 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 0.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.2 | 0.6 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.7 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 1.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.5 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.5 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.3 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.3 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.2 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.1 | 0.6 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 0.2 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 0.5 | GO:0070325 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.1 | 0.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.7 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.4 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.1 | 0.3 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 0.3 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.1 | 0.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 0.4 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0004953 | icosanoid receptor activity(GO:0004953) |
| 0.0 | 0.2 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.0 | 0.3 | GO:0004691 | AMP-activated protein kinase activity(GO:0004679) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.7 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.2 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 0.7 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0015198 | oligopeptide transporter activity(GO:0015198) oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.4 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.3 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0048270 | methionine adenosyltransferase regulator activity(GO:0048270) |
| 0.0 | 0.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.4 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 2.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.0 | 0.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.0 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.0 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.0 | 0.0 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.7 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.5 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.1 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 1.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 2.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 1.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.0 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.0 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |