Project
DANIO-CODE
Navigation
Downloads
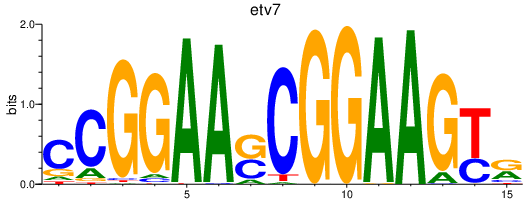
Results for etv7
Z-value: 1.00
Transcription factors associated with etv7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
etv7
|
ENSDARG00000089434 | ETS variant transcription factor 7 |
Activity profile of etv7 motif
Sorted Z-values of etv7 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of etv7
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_58523645 | 1.48 |
|
|
|
| chr14_-_10311891 | 1.45 |
ENSDART00000136649
|
si:dkey-92i17.2
|
si:dkey-92i17.2 |
| chr23_+_32175042 | 1.28 |
ENSDART00000138849
|
ENSDARG00000000887
|
ENSDARG00000000887 |
| chr19_+_348521 | 1.10 |
ENSDART00000114284
|
mcl1a
|
MCL1, BCL2 family apoptosis regulator a |
| chr23_+_32174669 | 1.02 |
ENSDART00000000992
|
ENSDARG00000000887
|
ENSDARG00000000887 |
| chr6_-_31377513 | 1.01 |
ENSDART00000145715
|
ak4
|
adenylate kinase 4 |
| chr13_+_49436597 | 1.01 |
ENSDART00000037559
ENSDART00000168799 ENSDART00000097845 |
ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr11_+_3940085 | 0.94 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr2_+_24851910 | 0.93 |
ENSDART00000133109
|
rps28
|
ribosomal protein S28 |
| chr15_+_37644266 | 0.92 |
ENSDART00000099456
|
psenen
|
presenilin enhancer gamma secretase subunit |
| chr5_+_27897765 | 0.91 |
ENSDART00000166737
ENSDART00000164418 |
nfr
|
notochord formation related |
| chr7_+_45747622 | 0.90 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr14_-_10311677 | 0.89 |
ENSDART00000133723
|
si:dkey-92i17.2
|
si:dkey-92i17.2 |
| chr7_+_45747395 | 0.89 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr8_-_31044580 | 0.87 |
ENSDART00000109885
|
snrnp200
|
small nuclear ribonucleoprotein 200 (U5) |
| chr11_+_3939876 | 0.82 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr8_+_4747546 | 0.82 |
ENSDART00000045533
|
tmem127
|
transmembrane protein 127 |
| chr8_+_17148864 | 0.79 |
ENSDART00000140531
|
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr3_+_61903223 | 0.78 |
ENSDART00000108945
|
GID4
|
GID complex subunit 4 homolog |
| chr13_-_33348231 | 0.78 |
ENSDART00000160520
|
mad2l1bp
|
MAD2L1 binding protein |
| chr3_+_61903372 | 0.78 |
ENSDART00000108945
|
GID4
|
GID complex subunit 4 homolog |
| chr9_-_22014861 | 0.73 |
ENSDART00000101986
|
mrpl30
|
mitochondrial ribosomal protein L30 |
| chr5_-_23525132 | 0.69 |
ENSDART00000051546
|
rps6ka3a
|
ribosomal protein S6 kinase a, polypeptide 3a |
| chr8_-_31044429 | 0.64 |
ENSDART00000109885
|
snrnp200
|
small nuclear ribonucleoprotein 200 (U5) |
| chr11_-_18174 | 0.62 |
ENSDART00000167814
|
prr13
|
proline rich 13 |
| chr16_+_7003624 | 0.62 |
ENSDART00000144763
|
eaf1
|
ELL associated factor 1 |
| chr14_-_10311764 | 0.62 |
ENSDART00000133723
|
si:dkey-92i17.2
|
si:dkey-92i17.2 |
| chr21_+_33418562 | 0.56 |
ENSDART00000053208
|
rps14
|
ribosomal protein S14 |
| chr16_-_52030972 | 0.56 |
ENSDART00000012055
|
rpl28
|
ribosomal protein L28 |
| chr22_-_20963723 | 0.55 |
|
|
|
| chr10_-_1669268 | 0.55 |
ENSDART00000125188
|
srsf9
|
serine/arginine-rich splicing factor 9 |
| chr1_+_39147577 | 0.54 |
ENSDART00000130270
|
irf2a
|
interferon regulatory factor 2a |
| chr14_+_50263527 | 0.53 |
ENSDART00000126260
|
anxa6
|
annexin A6 |
| chr16_+_41862759 | 0.53 |
|
|
|
| chr8_+_17148832 | 0.51 |
ENSDART00000140531
|
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr2_-_37832757 | 0.50 |
|
|
|
| chr2_+_24718602 | 0.49 |
ENSDART00000022379
|
nr2f6a
|
nuclear receptor subfamily 2, group F, member 6a |
| chr22_+_9493668 | 0.49 |
ENSDART00000143953
|
strip1
|
striatin interacting protein 1 |
| chr25_+_182037 | 0.49 |
ENSDART00000156469
|
RPS17
|
ribosomal protein S17 |
| chr4_+_2707833 | 0.48 |
ENSDART00000130623
|
dus4l
|
dihydrouridine synthase 4-like (S. cerevisiae) |
| chr8_+_388003 | 0.47 |
ENSDART00000067668
|
pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr20_-_37918013 | 0.47 |
ENSDART00000032978
|
nsl1
|
NSL1, MIS12 kinetochore complex component |
| chr6_+_60130210 | 0.46 |
ENSDART00000148557
|
aurka
|
aurora kinase A |
| chr2_+_24718905 | 0.46 |
ENSDART00000133818
|
nr2f6a
|
nuclear receptor subfamily 2, group F, member 6a |
| chr18_-_5619309 | 0.45 |
ENSDART00000177705
|
CABZ01081638.1
|
ENSDARG00000107008 |
| chr16_+_13970110 | 0.43 |
ENSDART00000121998
|
grwd1
|
glutamate-rich WD repeat containing 1 |
| chr8_+_387973 | 0.43 |
ENSDART00000067668
|
pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr8_+_41446054 | 0.43 |
ENSDART00000061144
|
arpc5la
|
actin related protein 2/3 complex, subunit 5-like, a |
| chr8_+_387564 | 0.42 |
ENSDART00000167361
|
pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr9_-_22014768 | 0.42 |
ENSDART00000101986
|
mrpl30
|
mitochondrial ribosomal protein L30 |
| chr7_-_45747112 | 0.42 |
ENSDART00000172591
|
zgc:162297
|
zgc:162297 |
| chr17_+_43749940 | 0.42 |
ENSDART00000133317
ENSDART00000140316 ENSDART00000142929 ENSDART00000133874 |
zgc:66313
|
zgc:66313 |
| chr8_+_1122000 | 0.42 |
ENSDART00000127252
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr1_-_358810 | 0.41 |
ENSDART00000098627
|
pros1
|
protein S (alpha) |
| chr25_+_446920 | 0.41 |
ENSDART00000104717
|
rsl24d1
|
ribosomal L24 domain containing 1 |
| chr16_-_9562608 | 0.41 |
ENSDART00000126154
|
prpf3
|
PRP3 pre-mRNA processing factor 3 homolog (yeast) |
| chr7_+_26274744 | 0.41 |
ENSDART00000135313
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr6_+_43018443 | 0.39 |
ENSDART00000064888
|
tcta
|
T-cell leukemia translocation altered gene |
| chr10_+_17790406 | 0.39 |
ENSDART00000135044
|
pigo
|
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr21_-_13026036 | 0.39 |
ENSDART00000135623
|
fam219aa
|
family with sequence similarity 219, member Aa |
| chr23_-_6793566 | 0.38 |
ENSDART00000171714
|
baz2a
|
bromodomain adjacent to zinc finger domain, 2A |
| chr18_-_40893823 | 0.38 |
ENSDART00000059194
|
snrpd2
|
small nuclear ribonucleoprotein D2 polypeptide |
| chr19_-_37921386 | 0.38 |
ENSDART00000018255
|
ilf2
|
interleukin enhancer binding factor 2 |
| chr3_+_24381364 | 0.37 |
ENSDART00000133898
|
dnal4a
|
dynein, axonemal, light chain 4a |
| chr1_+_54212090 | 0.37 |
ENSDART00000140375
|
zfyve27
|
zinc finger, FYVE domain containing 27 |
| chr4_+_1750689 | 0.37 |
ENSDART00000146779
|
slc38a2
|
solute carrier family 38, member 2 |
| chr16_+_55121654 | 0.37 |
ENSDART00000149449
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr15_-_34709255 | 0.36 |
ENSDART00000099792
|
sostdc1a
|
sclerostin domain containing 1a |
| chr6_-_22910388 | 0.35 |
ENSDART00000164114
|
grb2a
|
growth factor receptor-bound protein 2a |
| chr4_-_16611960 | 0.35 |
|
|
|
| chr18_+_15809534 | 0.34 |
ENSDART00000014188
|
ube2na
|
ubiquitin-conjugating enzyme E2Na |
| chr21_-_36768113 | 0.34 |
ENSDART00000018350
|
cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr2_+_36626618 | 0.33 |
ENSDART00000159541
|
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr3_-_15905495 | 0.32 |
ENSDART00000023859
|
atp6v0ca
|
ATPase, H+ transporting, lysosomal, V0 subunit ca |
| chr14_+_10352087 | 0.32 |
|
|
|
| chr14_+_51692872 | 0.32 |
ENSDART00000163856
|
noa1
|
nitric oxide associated 1 |
| chr22_-_6164703 | 0.32 |
ENSDART00000129829
|
si:rp71-36a1.5
|
si:rp71-36a1.5 |
| chr16_+_7003851 | 0.31 |
ENSDART00000145141
|
eaf1
|
ELL associated factor 1 |
| chr2_-_24661477 | 0.31 |
ENSDART00000078975
ENSDART00000155677 |
trnau1apb
|
tRNA selenocysteine 1 associated protein 1b |
| chr7_+_49628680 | 0.30 |
ENSDART00000007487
|
apip
|
APAF1 interacting protein |
| chr15_+_44909293 | 0.30 |
ENSDART00000111373
|
pdgfd
|
platelet derived growth factor d |
| chr22_+_120930 | 0.30 |
ENSDART00000114777
|
nckipsd
|
NCK interacting protein with SH3 domain |
| chr25_+_181958 | 0.29 |
ENSDART00000155412
|
RPS17
|
ribosomal protein S17 |
| chr7_+_26274687 | 0.29 |
ENSDART00000135313
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr15_-_5911817 | 0.29 |
|
|
|
| chr24_+_29278310 | 0.29 |
ENSDART00000101641
|
prmt6
|
protein arginine methyltransferase 6 |
| chr16_-_9562744 | 0.28 |
ENSDART00000126154
|
prpf3
|
PRP3 pre-mRNA processing factor 3 homolog (yeast) |
| chr13_+_9227395 | 0.28 |
ENSDART00000047740
|
wdr32
|
WD repeat domain 32 |
| chr22_+_223797 | 0.27 |
|
|
|
| chr3_-_25361708 | 0.27 |
ENSDART00000147322
|
grb2b
|
growth factor receptor-bound protein 2b |
| chr24_+_33548367 | 0.27 |
ENSDART00000166666
|
zgc:101800
|
zgc:101800 |
| chr8_+_52544578 | 0.26 |
ENSDART00000127729
|
stambpb
|
STAM binding protein b |
| chr2_-_40067342 | 0.26 |
|
|
|
| chr15_+_44909356 | 0.26 |
ENSDART00000111373
|
pdgfd
|
platelet derived growth factor d |
| chr3_-_25361866 | 0.26 |
ENSDART00000147322
|
grb2b
|
growth factor receptor-bound protein 2b |
| chr8_+_7815256 | 0.26 |
ENSDART00000165575
|
cxxc1a
|
CXXC finger protein 1a |
| chr14_+_50232242 | 0.26 |
ENSDART00000110099
|
rmnd5b
|
required for meiotic nuclear division 5 homolog B |
| KN150139v1_+_12765 | 0.26 |
|
|
|
| chr16_-_52031031 | 0.25 |
ENSDART00000012055
|
rpl28
|
ribosomal protein L28 |
| chr7_-_45747162 | 0.25 |
ENSDART00000172591
|
zgc:162297
|
zgc:162297 |
| chr22_-_120910 | 0.25 |
|
|
|
| chr25_-_13562558 | 0.24 |
ENSDART00000045488
|
csnk2a2b
|
casein kinase 2, alpha prime polypeptide b |
| chr15_-_5912105 | 0.24 |
|
|
|
| chr2_-_57827134 | 0.23 |
ENSDART00000138307
|
si:dkeyp-68b7.5
|
si:dkeyp-68b7.5 |
| chr13_+_6060600 | 0.23 |
ENSDART00000159517
|
ppm1g
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr9_-_33518818 | 0.23 |
ENSDART00000140039
|
rpl8
|
ribosomal protein L8 |
| chr2_+_35871754 | 0.23 |
ENSDART00000134918
|
dhx9
|
DEAH (Asp-Glu-Ala-His) box helicase 9 |
| chr3_+_17783319 | 0.23 |
ENSDART00000104299
|
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr2_+_54665352 | 0.22 |
|
|
|
| chr25_+_16784585 | 0.22 |
ENSDART00000020259
|
zgc:77158
|
zgc:77158 |
| chr20_-_4750304 | 0.22 |
ENSDART00000053870
|
galca
|
galactosylceramidase a |
| chr1_+_10423544 | 0.22 |
ENSDART00000109858
|
knstrn
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr17_-_51032797 | 0.22 |
ENSDART00000130412
ENSDART00000123746 ENSDART00000162717 |
aqr
|
aquarius intron-binding spliceosomal factor |
| chr7_-_33080261 | 0.22 |
ENSDART00000114041
|
anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr11_-_14943957 | 0.22 |
ENSDART00000162938
|
hug
|
HuG |
| chr20_+_39441958 | 0.21 |
ENSDART00000009164
|
esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr21_+_7868319 | 0.21 |
ENSDART00000121813
ENSDART00000021272 |
wdr41
|
WD repeat domain 41 |
| chr1_-_16668277 | 0.21 |
ENSDART00000135012
|
BX511093.1
|
ENSDARG00000092109 |
| chr14_+_11872906 | 0.20 |
ENSDART00000054626
|
hdac3
|
histone deacetylase 3 |
| chr11_-_6858626 | 0.20 |
ENSDART00000168372
|
lsm4
|
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr11_+_5746030 | 0.19 |
ENSDART00000179139
|
arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr1_-_54089140 | 0.19 |
|
|
|
| chr21_-_30217568 | 0.18 |
ENSDART00000101054
|
dnajc18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr13_+_9227690 | 0.17 |
ENSDART00000047740
|
wdr32
|
WD repeat domain 32 |
| chr13_-_5849887 | 0.17 |
ENSDART00000160823
|
actr2b
|
ARP2 actin related protein 2b homolog |
| chr13_-_3191411 | 0.17 |
ENSDART00000114040
|
ubr2
|
ubiquitin protein ligase E3 component n-recognin 2 |
| chr5_-_68712851 | 0.17 |
ENSDART00000109487
|
CABZ01032476.1
|
ENSDARG00000074645 |
| chr22_-_9832520 | 0.17 |
|
|
|
| chr14_-_51692654 | 0.16 |
ENSDART00000162719
|
polr2b
|
polymerase (RNA) II (DNA directed) polypeptide B |
| chr19_+_43090603 | 0.16 |
ENSDART00000018328
|
fbxl2
|
F-box and leucine-rich repeat protein 2 |
| chr22_-_469154 | 0.16 |
ENSDART00000106645
ENSDART00000067637 |
dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr25_-_13562597 | 0.16 |
ENSDART00000045488
|
csnk2a2b
|
casein kinase 2, alpha prime polypeptide b |
| chr8_-_11508437 | 0.16 |
ENSDART00000081909
|
si:ch211-248e11.2
|
si:ch211-248e11.2 |
| chr19_+_43090664 | 0.16 |
ENSDART00000018328
|
fbxl2
|
F-box and leucine-rich repeat protein 2 |
| chr3_+_15294683 | 0.16 |
|
|
|
| chr2_+_24852094 | 0.15 |
ENSDART00000052063
|
rps28
|
ribosomal protein S28 |
| chr22_+_1430747 | 0.15 |
|
|
|
| chr13_+_6060769 | 0.15 |
ENSDART00000159517
|
ppm1g
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr22_-_9832436 | 0.15 |
|
|
|
| chr13_-_28133363 | 0.15 |
ENSDART00000041036
|
smap1
|
small ArfGAP 1 |
| chr12_+_10078043 | 0.15 |
ENSDART00000152369
|
si:dkeyp-118b1.2
|
si:dkeyp-118b1.2 |
| chr19_+_40792676 | 0.15 |
ENSDART00000110699
|
vps50
|
VPS50 EARP/GARPII complex subunit |
| chr7_-_25777968 | 0.15 |
ENSDART00000101124
|
rnaseka
|
ribonuclease, RNase K a |
| chr4_-_9666436 | 0.15 |
ENSDART00000133214
|
dmtf1
|
cyclin D binding myb-like transcription factor 1 |
| chr6_+_31377287 | 0.14 |
|
|
|
| chr2_+_57894141 | 0.14 |
ENSDART00000171264
ENSDART00000163278 |
si:ch211-155e24.3
|
si:ch211-155e24.3 |
| chr1_+_966385 | 0.14 |
ENSDART00000051919
|
n6amt1
|
N-6 adenine-specific DNA methyltransferase 1 (putative) |
| chr18_+_15809437 | 0.14 |
ENSDART00000014188
|
ube2na
|
ubiquitin-conjugating enzyme E2Na |
| chr8_-_6875110 | 0.13 |
ENSDART00000014915
|
asb6
|
ankyrin repeat and SOCS box containing 6 |
| chr22_-_111800 | 0.12 |
ENSDART00000163198
ENSDART00000168678 |
capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr6_+_49724271 | 0.12 |
ENSDART00000002693
|
stx16
|
syntaxin 16 |
| chr1_+_50763649 | 0.12 |
ENSDART00000074294
|
actr2a
|
ARP2 actin related protein 2a homolog |
| chr8_-_17148743 | 0.12 |
ENSDART00000025803
|
pola2
|
polymerase (DNA directed), alpha 2 |
| chr17_-_43749763 | 0.12 |
ENSDART00000167422
|
ahsa1b
|
AHA1, activator of heat shock protein ATPase homolog 1b |
| chr20_-_4750503 | 0.11 |
ENSDART00000053870
|
galca
|
galactosylceramidase a |
| chr3_+_36830725 | 0.11 |
ENSDART00000150917
|
ENSDARG00000070174
|
ENSDARG00000070174 |
| chr16_+_9835124 | 0.11 |
ENSDART00000020859
|
pip5k1ab
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, b |
| chr11_-_16840339 | 0.11 |
ENSDART00000122222
|
suclg2
|
succinate-CoA ligase, GDP-forming, beta subunit |
| chr14_-_38538068 | 0.11 |
ENSDART00000173300
|
uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr4_-_16611908 | 0.11 |
|
|
|
| chr10_+_17503275 | 0.11 |
|
|
|
| chr1_+_499101 | 0.10 |
ENSDART00000108579
|
blzf1
|
basic leucine zipper nuclear factor 1 |
| chr25_-_36767376 | 0.10 |
ENSDART00000073439
|
trim44
|
tripartite motif containing 44 |
| chr20_+_37918171 | 0.10 |
ENSDART00000064692
|
tatdn3
|
TatD DNase domain containing 3 |
| chr2_+_37833161 | 0.10 |
ENSDART00000166352
|
sdr39u1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr22_+_1436176 | 0.10 |
|
|
|
| chr8_-_31044627 | 0.10 |
ENSDART00000109885
|
snrnp200
|
small nuclear ribonucleoprotein 200 (U5) |
| chr1_+_10423372 | 0.09 |
ENSDART00000109858
|
knstrn
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr7_+_26274252 | 0.09 |
ENSDART00000164824
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr5_-_45294605 | 0.08 |
ENSDART00000097648
ENSDART00000142782 |
crfb6
|
cytokine receptor family member b6 |
| chr4_-_2707563 | 0.08 |
ENSDART00000021953
ENSDART00000150344 |
cog5
|
component of oligomeric golgi complex 5 |
| chr18_-_11439216 | 0.08 |
|
|
|
| chr1_-_10423289 | 0.08 |
|
|
|
| chr8_+_7815304 | 0.08 |
ENSDART00000165575
|
cxxc1a
|
CXXC finger protein 1a |
| chr5_-_68712571 | 0.08 |
ENSDART00000109487
|
CABZ01032476.1
|
ENSDARG00000074645 |
| chr1_+_58608185 | 0.07 |
ENSDART00000161872
ENSDART00000160658 |
parn
|
poly(A)-specific ribonuclease (deadenylation nuclease) |
| chr7_+_25778546 | 0.07 |
ENSDART00000173611
|
si:dkey-6n21.12
|
si:dkey-6n21.12 |
| chr14_+_11873110 | 0.07 |
ENSDART00000171002
|
hdac3
|
histone deacetylase 3 |
| chr5_-_3964337 | 0.07 |
ENSDART00000168096
|
srrt
|
serrate RNA effector molecule homolog (Arabidopsis) |
| chr20_+_4750766 | 0.07 |
ENSDART00000153486
|
lgals8a
|
galectin 8a |
| chr21_-_2147438 | 0.07 |
ENSDART00000172276
|
zgc:163077
|
zgc:163077 |
| chr7_+_20046382 | 0.06 |
ENSDART00000023089
|
acadvl
|
acyl-CoA dehydrogenase, very long chain |
| chr13_+_5958225 | 0.06 |
ENSDART00000049328
|
fam120b
|
family with sequence similarity 120B |
| chr19_+_27895558 | 0.05 |
ENSDART00000103922
|
atat1
|
alpha tubulin acetyltransferase 1 |
| chr19_+_6985574 | 0.05 |
ENSDART00000151502
|
si:ch1073-127d16.1
|
si:ch1073-127d16.1 |
| chr24_-_16993914 | 0.05 |
|
|
|
| chr15_-_26998580 | 0.04 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| chr22_-_342682 | 0.03 |
ENSDART00000067633
|
necap2
|
NECAP endocytosis associated 2 |
| chr6_+_60130111 | 0.03 |
ENSDART00000148557
|
aurka
|
aurora kinase A |
| chr6_+_49724436 | 0.03 |
ENSDART00000154738
|
stx16
|
syntaxin 16 |
| chr3_+_27534273 | 0.02 |
ENSDART00000103660
|
clcn7
|
chloride channel 7 |
| chr7_+_28906162 | 0.02 |
ENSDART00000008096
|
aph1b
|
APH1B gamma secretase subunit |
| chr25_-_18850809 | 0.02 |
ENSDART00000091549
|
nt5dc3
|
5'-nucleotidase domain containing 3 |
| chr4_-_5362973 | 0.02 |
ENSDART00000150697
|
si:dkey-14d8.1
|
si:dkey-14d8.1 |
| chr19_+_7576862 | 0.01 |
ENSDART00000010862
|
mrpl24
|
mitochondrial ribosomal protein L24 |
| chr16_+_33976740 | 0.01 |
ENSDART00000167240
|
snip1
|
Smad nuclear interacting protein |
| chr6_+_94581 | 0.01 |
ENSDART00000125176
|
ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr23_+_20718153 | 0.00 |
ENSDART00000133131
|
uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr5_-_907020 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0000388 | spliceosome conformational change to release U4 (or U4atac) and U1 (or U11)(GO:0000388) |
| 0.4 | 1.3 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
| 0.2 | 0.9 | GO:0042987 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.2 | 1.0 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.2 | 2.5 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.4 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.5 | GO:0003413 | plasma membrane repair(GO:0001778) chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.1 | 0.3 | GO:0006683 | galactosylceramide catabolic process(GO:0006683) galactolipid catabolic process(GO:0019376) |
| 0.1 | 0.3 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.1 | 0.5 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 0.4 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 1.1 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 | 1.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.4 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.3 | GO:2001032 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 0.6 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.5 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.3 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.1 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.4 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 1.3 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.5 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.4 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 0.7 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.0 | 0.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 1.8 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.0 | 0.3 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 1.0 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.7 | GO:0043588 | skin development(GO:0043588) |
| 0.0 | 0.8 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.2 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.1 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.0 | 0.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.1 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.0 | 0.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.3 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.4 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.3 | 1.8 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.5 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 1.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.5 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.7 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 1.1 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.1 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 2.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.6 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.7 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 1.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.5 | GO:0015934 | large ribosomal subunit(GO:0015934) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 0.5 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 0.6 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.4 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.1 | 0.3 | GO:0004336 | galactosylceramidase activity(GO:0004336) |
| 0.1 | 1.0 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.5 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.3 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 0.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.4 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.4 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.7 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 1.0 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.4 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 1.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 1.8 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 3.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 1.0 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.2 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.0 | 0.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.1 | GO:0042803 | protein homodimerization activity(GO:0042803) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.8 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 2.2 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |