Project
DANIO-CODE
Navigation
Downloads
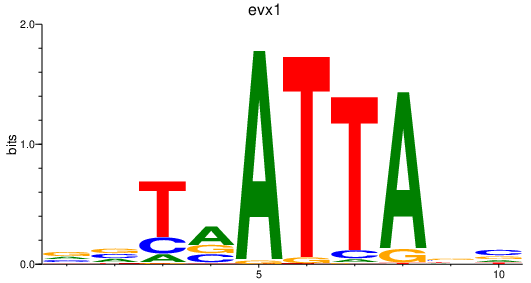
Results for evx1
Z-value: 0.63
Transcription factors associated with evx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
evx1
|
ENSDARG00000099365 | even-skipped homeobox 1 |
|
evx1
|
ENSDARG00000100087 | even-skipped homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| evx1 | dr10_dc_chr19_-_20148533_20148589 | 0.84 | 5.2e-05 | Click! |
Activity profile of evx1 motif
Sorted Z-values of evx1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of evx1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_25623974 | 1.88 |
ENSDART00000173602
|
cd99l2
|
CD99 molecule-like 2 |
| chr6_+_56157608 | 1.85 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr8_+_31426328 | 1.72 |
ENSDART00000135101
|
sepp1a
|
selenoprotein P, plasma, 1a |
| chr7_+_30516734 | 1.65 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr25_-_13737344 | 1.59 |
|
|
|
| chr22_-_26333957 | 1.45 |
ENSDART00000130493
|
capn2b
|
calpain 2, (m/II) large subunit b |
| chr1_+_26330186 | 1.38 |
ENSDART00000102322
|
bnc2
|
basonuclin 2 |
| chr17_+_135165 | 1.27 |
ENSDART00000166339
|
klhdc2
|
kelch domain containing 2 |
| chr13_+_27102377 | 1.22 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr3_+_18249107 | 1.21 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr7_-_46510632 | 1.15 |
|
|
|
| chr9_+_13149212 | 1.11 |
ENSDART00000141005
|
fam117bb
|
family with sequence similarity 117, member Bb |
| chr15_-_1858350 | 1.11 |
ENSDART00000082026
|
mmp28
|
matrix metallopeptidase 28 |
| chr4_+_2215426 | 1.09 |
ENSDART00000168370
|
frs2a
|
fibroblast growth factor receptor substrate 2a |
| chr1_-_37990863 | 1.07 |
ENSDART00000132402
|
gpm6ab
|
glycoprotein M6Ab |
| chr2_+_29992879 | 1.06 |
ENSDART00000056748
|
en2b
|
engrailed homeobox 2b |
| chr20_-_40823307 | 1.04 |
ENSDART00000061261
|
cx43
|
connexin 43 |
| chr2_-_23516930 | 1.04 |
ENSDART00000165355
|
prrx1a
|
paired related homeobox 1a |
| chr17_-_31678247 | 1.02 |
ENSDART00000143090
|
lin52
|
lin-52 DREAM MuvB core complex component |
| chr12_+_20230575 | 1.02 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr24_+_7832020 | 0.99 |
ENSDART00000019705
|
bmp6
|
bone morphogenetic protein 6 |
| chr7_-_52848084 | 0.97 |
ENSDART00000172179
ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr4_+_9177011 | 0.96 |
ENSDART00000057254
|
nfyba
|
nuclear transcription factor Y, beta a |
| chr19_+_31046291 | 0.96 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr9_-_20562496 | 0.95 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr2_-_17564676 | 0.94 |
ENSDART00000144251
|
artnb
|
artemin b |
| chr17_+_134921 | 0.92 |
ENSDART00000166339
|
klhdc2
|
kelch domain containing 2 |
| chr13_+_27102308 | 0.92 |
ENSDART00000145901
|
rin2
|
Ras and Rab interactor 2 |
| chr3_+_17387551 | 0.91 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr16_+_43249142 | 0.90 |
ENSDART00000154493
|
adam22
|
ADAM metallopeptidase domain 22 |
| chr5_-_71340996 | 0.90 |
ENSDART00000162526
|
smyd1a
|
SET and MYND domain containing 1a |
| chr11_-_6442836 | 0.88 |
ENSDART00000004483
|
zgc:162969
|
zgc:162969 |
| chr5_-_13872895 | 0.88 |
ENSDART00000164698
|
CT009570.1
|
ENSDARG00000104490 |
| chr8_-_15091823 | 0.85 |
ENSDART00000142358
|
bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr5_-_33156615 | 0.85 |
ENSDART00000159058
|
dab2ipb
|
DAB2 interacting protein b |
| chr7_+_65532390 | 0.85 |
ENSDART00000156683
|
CT573494.2
|
ENSDARG00000097673 |
| chr2_-_23517033 | 0.85 |
ENSDART00000041365
|
prrx1a
|
paired related homeobox 1a |
| chr11_-_40414239 | 0.78 |
ENSDART00000165163
|
si:ch211-222l21.1
|
si:ch211-222l21.1 |
| chr9_-_20562293 | 0.75 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr7_+_55883471 | 0.75 |
ENSDART00000073594
|
ankrd11
|
ankyrin repeat domain 11 |
| chr22_+_16282153 | 0.75 |
ENSDART00000162685
ENSDART00000105678 |
lrrc39
|
leucine rich repeat containing 39 |
| chr5_+_68410884 | 0.74 |
ENSDART00000153691
|
CU928129.1
|
ENSDARG00000097815 |
| chr9_-_19154264 | 0.71 |
ENSDART00000081878
|
pou1f1
|
POU class 1 homeobox 1 |
| chr7_-_56493164 | 0.70 |
|
|
|
| chr16_+_29060022 | 0.68 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr17_-_25630666 | 0.67 |
ENSDART00000149060
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr22_-_864745 | 0.67 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr7_-_23775835 | 0.66 |
ENSDART00000144616
|
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr24_+_40292617 | 0.65 |
|
|
|
| chr16_-_54634679 | 0.63 |
ENSDART00000075275
ENSDART00000115024 |
pklr
|
pyruvate kinase, liver and RBC |
| chr20_-_53271601 | 0.63 |
|
|
|
| chr10_-_17630376 | 0.63 |
ENSDART00000113101
|
smarcad1b
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 b |
| chr17_+_24668907 | 0.63 |
ENSDART00000034263
ENSDART00000135794 |
sepn1
|
selenoprotein N, 1 |
| chr20_-_23526954 | 0.62 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr23_-_28286971 | 0.62 |
|
|
|
| chr7_-_72278552 | 0.61 |
ENSDART00000168532
|
HECTD4
|
HECT domain E3 ubiquitin protein ligase 4 |
| chr6_+_24299180 | 0.61 |
ENSDART00000167482
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr13_-_49695322 | 0.60 |
|
|
|
| chr6_-_6330710 | 0.59 |
ENSDART00000137642
|
cep170ab
|
centrosomal protein 170Ab |
| chr11_-_42626842 | 0.59 |
ENSDART00000052912
|
pcdh20
|
protocadherin 20 |
| chr23_+_44137593 | 0.56 |
ENSDART00000148470
|
CU993818.1
|
ENSDARG00000095873 |
| chr13_+_9216834 | 0.56 |
ENSDART00000122695
|
pomk
|
protein-O-mannose kinase |
| chr6_-_39346614 | 0.55 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr6_-_8075384 | 0.54 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr25_-_1989785 | 0.52 |
ENSDART00000156925
|
wnt7bb
|
wingless-type MMTV integration site family, member 7Bb |
| chr17_+_135261 | 0.51 |
ENSDART00000166339
|
klhdc2
|
kelch domain containing 2 |
| chr16_+_11138879 | 0.51 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr18_-_43890514 | 0.51 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr16_+_11138924 | 0.50 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr25_+_35855292 | 0.49 |
ENSDART00000152801
|
ankrd27
|
ankyrin repeat domain 27 (VPS9 domain) |
| chr23_+_31986806 | 0.48 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr8_-_37217197 | 0.48 |
ENSDART00000178556
|
rbm39b
|
RNA binding motif protein 39b |
| chr1_+_26330335 | 0.47 |
ENSDART00000102322
|
bnc2
|
basonuclin 2 |
| KN149830v1_-_23361 | 0.44 |
|
|
|
| chr5_+_64050769 | 0.44 |
ENSDART00000073950
|
olfm1a
|
olfactomedin 1a |
| chr7_-_25624212 | 0.44 |
ENSDART00000173505
|
cd99l2
|
CD99 molecule-like 2 |
| chr21_+_26690161 | 0.44 |
ENSDART00000065392
|
calm3b
|
calmodulin 3b (phosphorylase kinase, delta) |
| chr7_-_39266252 | 0.43 |
ENSDART00000173965
|
otog
|
otogelin |
| chr25_+_5922190 | 0.43 |
ENSDART00000128816
|
sv2
|
synaptic vesicle glycoprotein 2 |
| chr15_-_18130992 | 0.42 |
ENSDART00000113142
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr4_+_11385828 | 0.41 |
|
|
|
| chr5_-_71277652 | 0.41 |
|
|
|
| chr9_+_36316158 | 0.40 |
ENSDART00000176763
|
lrp1bb
|
low density lipoprotein receptor-related protein 1Bb |
| chr20_-_46458839 | 0.40 |
ENSDART00000153087
|
bmf2
|
Bcl2 modifying factor 2 |
| chr1_-_37990935 | 0.39 |
ENSDART00000132402
|
gpm6ab
|
glycoprotein M6Ab |
| chr16_+_35708856 | 0.38 |
ENSDART00000161393
|
map7d1a
|
MAP7 domain containing 1a |
| chr13_+_32987105 | 0.37 |
ENSDART00000085719
|
si:ch211-10a23.2
|
si:ch211-10a23.2 |
| chr11_-_40414516 | 0.36 |
ENSDART00000165163
|
si:ch211-222l21.1
|
si:ch211-222l21.1 |
| chr24_-_24859334 | 0.36 |
ENSDART00000080997
|
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr21_-_26453406 | 0.36 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr14_-_33704021 | 0.35 |
ENSDART00000149396
ENSDART00000123607 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr15_+_880573 | 0.33 |
ENSDART00000115077
|
zgc:174573
|
zgc:174573 |
| chr13_-_45338347 | 0.33 |
ENSDART00000043345
|
rsrp1
|
arginine/serine-rich protein 1 |
| chr2_-_23516991 | 0.32 |
ENSDART00000165355
|
prrx1a
|
paired related homeobox 1a |
| chr3_-_55278137 | 0.32 |
ENSDART00000123544
|
tex2
|
testis expressed 2 |
| chr16_+_17481157 | 0.32 |
ENSDART00000173448
|
fam131bb
|
family with sequence similarity 131, member Bb |
| chr9_-_39190534 | 0.32 |
|
|
|
| chr6_-_60088120 | 0.31 |
|
|
|
| chr7_-_25624128 | 0.31 |
ENSDART00000173505
|
cd99l2
|
CD99 molecule-like 2 |
| chr22_-_11106940 | 0.30 |
ENSDART00000016873
|
atp6ap2
|
ATPase, H+ transporting, lysosomal accessory protein 2 |
| chr2_-_32842855 | 0.30 |
ENSDART00000098834
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr3_-_30948297 | 0.30 |
ENSDART00000145636
|
ELOB (1 of many)
|
elongin B |
| chr11_+_38013238 | 0.29 |
ENSDART00000171496
|
CDK18
|
cyclin dependent kinase 18 |
| chr8_+_36888422 | 0.29 |
|
|
|
| chr16_+_43249077 | 0.29 |
ENSDART00000154493
|
adam22
|
ADAM metallopeptidase domain 22 |
| chr17_-_23426752 | 0.28 |
ENSDART00000104715
|
pcgf5a
|
polycomb group ring finger 5a |
| chr14_-_6831308 | 0.28 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr23_-_12310778 | 0.26 |
ENSDART00000131256
|
phactr3a
|
phosphatase and actin regulator 3a |
| chr1_+_50547214 | 0.26 |
ENSDART00000132244
|
btbd3a
|
BTB (POZ) domain containing 3a |
| chr23_+_34037058 | 0.26 |
ENSDART00000140666
|
prosc
|
pyridoxal phosphate binding protein |
| chr17_+_37280302 | 0.25 |
ENSDART00000128879
|
hadhab
|
hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase (trifunctional protein), alpha subunit b |
| chr8_+_2380040 | 0.24 |
ENSDART00000141263
|
ENKD1
|
enkurin domain containing 1 |
| chr7_-_64172974 | 0.24 |
ENSDART00000172619
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr4_+_12613575 | 0.23 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr20_-_53271658 | 0.23 |
|
|
|
| chr19_+_9113669 | 0.22 |
|
|
|
| chr15_-_16241341 | 0.22 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr7_+_65532771 | 0.21 |
ENSDART00000082740
|
CT573494.2
|
ENSDARG00000097673 |
| chr7_+_65532871 | 0.21 |
ENSDART00000082740
|
CT573494.2
|
ENSDARG00000097673 |
| chr20_+_54544931 | 0.21 |
|
|
|
| chr24_-_39630728 | 0.20 |
ENSDART00000031486
|
lyrm1
|
LYR motif containing 1 |
| chr25_-_5921772 | 0.20 |
ENSDART00000075184
|
snx1a
|
sorting nexin 1a |
| chr17_-_135071 | 0.20 |
|
|
|
| chr24_+_16402613 | 0.20 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr1_-_10373506 | 0.20 |
|
|
|
| chr17_-_21180028 | 0.19 |
ENSDART00000104708
|
abhd12
|
abhydrolase domain containing 12 |
| chr2_-_23516868 | 0.17 |
ENSDART00000165355
|
prrx1a
|
paired related homeobox 1a |
| chr3_+_32700750 | 0.16 |
ENSDART00000139410
|
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr3_-_55278055 | 0.16 |
ENSDART00000123544
|
tex2
|
testis expressed 2 |
| chr20_+_26012369 | 0.15 |
ENSDART00000146004
|
ttbk2b
|
tau tubulin kinase 2b |
| chr1_+_32915075 | 0.15 |
ENSDART00000114384
|
znf654
|
zinc finger protein 654 |
| chr21_+_11151424 | 0.15 |
ENSDART00000163432
|
arid6
|
AT-rich interaction domain 6 |
| chr20_-_23527004 | 0.14 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr23_-_19577146 | 0.13 |
ENSDART00000143288
|
asb14b
|
ankyrin repeat and SOCS box containing 14b |
| chr25_-_35530474 | 0.13 |
|
|
|
| chr20_+_26441985 | 0.12 |
|
|
|
| chr5_-_23650217 | 0.12 |
ENSDART00000080609
|
mrps31
|
mitochondrial ribosomal protein S31 |
| chr11_-_40414664 | 0.12 |
ENSDART00000165163
|
si:ch211-222l21.1
|
si:ch211-222l21.1 |
| chr2_-_27065203 | 0.11 |
ENSDART00000148110
|
si:dkey-181m9.8
|
si:dkey-181m9.8 |
| chr6_-_16328987 | 0.10 |
ENSDART00000083305
|
slc19a2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr14_-_3951077 | 0.10 |
|
|
|
| chr14_-_1187074 | 0.10 |
ENSDART00000106672
|
arl9
|
ADP-ribosylation factor-like 9 |
| chr3_+_27655753 | 0.09 |
ENSDART00000086994
|
nat15
|
N-acetyltransferase 15 (GCN5-related, putative) |
| chr2_-_22173254 | 0.09 |
ENSDART00000137538
|
rab2a
|
RAB2A, member RAS oncogene family |
| chr4_+_11465367 | 0.08 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
| chr3_-_29932835 | 0.06 |
|
|
|
| chr2_-_32842678 | 0.06 |
ENSDART00000098834
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr17_-_10684072 | 0.05 |
|
|
|
| chr8_-_31098193 | 0.04 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr6_+_55203104 | 0.04 |
ENSDART00000083668
|
manbal
|
mannosidase beta like |
| chr22_+_2074020 | 0.03 |
ENSDART00000106540
|
znf1161
|
zinc finger protein 1161 |
| chr13_-_21529695 | 0.02 |
ENSDART00000100925
|
mxtx1
|
mix-type homeobox gene 1 |
| chr6_-_16156294 | 0.02 |
ENSDART00000160809
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr23_+_7614867 | 0.02 |
ENSDART00000011554
|
tm9sf4
|
transmembrane 9 superfamily protein member 4 |
| chr15_-_5911817 | 0.02 |
|
|
|
| chr10_-_2944221 | 0.01 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| KN150030v1_-_22642 | 0.00 |
ENSDART00000175410
|
CABZ01079802.1
|
ENSDARG00000106760 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0086065 | atrial cardiac muscle cell action potential(GO:0086014) cell-cell signaling involved in cardiac conduction(GO:0086019) atrial cardiac muscle cell to AV node cell signaling(GO:0086026) cell communication involved in cardiac conduction(GO:0086065) atrial cardiac muscle cell to AV node cell communication(GO:0086066) |
| 0.3 | 0.9 | GO:0001659 | temperature homeostasis(GO:0001659) sleep(GO:0030431) |
| 0.2 | 1.0 | GO:0003379 | establishment of cell polarity involved in gastrulation cell migration(GO:0003379) |
| 0.2 | 0.9 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.2 | 0.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.4 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 0.8 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 1.9 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.1 | 1.5 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 0.6 | GO:0036268 | swimming(GO:0036268) |
| 0.1 | 1.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 1.1 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 0.9 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 1.9 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.1 | 0.9 | GO:0030241 | skeletal muscle thin filament assembly(GO:0030240) skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.4 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.1 | 0.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.6 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 0.5 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.4 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 1.0 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.7 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 1.1 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.5 | GO:0015696 | ammonium transport(GO:0015696) |
| 0.0 | 0.2 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.3 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0045117 | azole transport(GO:0045117) |
| 0.0 | 0.6 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 0.5 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.6 | GO:0006096 | glycolytic process(GO:0006096) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 1.0 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 1.6 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.0 | GO:0031838 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.6 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.2 | 0.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 1.0 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 1.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.0 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.5 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.1 | 1.0 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 1.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.6 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 1.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.2 | GO:0008665 | tRNA 2'-phosphotransferase activity(GO:0000215) 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 2.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.9 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.1 | GO:0090482 | azole transporter activity(GO:0045118) vitamin transmembrane transporter activity(GO:0090482) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 3.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 1.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 2.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 0.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 0.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 1.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |