Project
DANIO-CODE
Navigation
Downloads
Results for figla
Z-value: 1.12
Transcription factors associated with figla
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
figla
|
ENSDARG00000087166 | folliculogenesis specific bHLH transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| figla | dr10_dc_chr5_-_13347964_13348049 | 0.84 | 4.3e-05 | Click! |
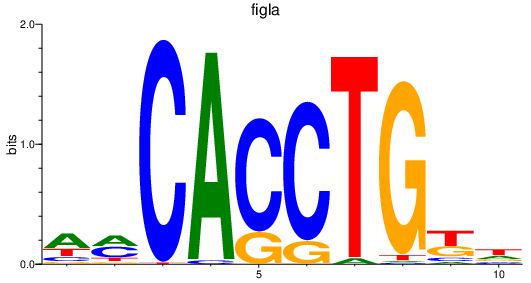
Activity profile of figla motif
Sorted Z-values of figla motif
Network of associatons between targets according to the STRING database.
First level regulatory network of figla
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_57254393 | 4.86 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr10_-_25246786 | 4.43 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr15_+_38397772 | 4.41 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr7_+_58448953 | 4.17 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr12_-_4264663 | 3.73 |
ENSDART00000152521
|
ca15b
|
carbonic anhydrase XVb |
| KN150456v1_-_19515 | 3.63 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr16_-_42116471 | 3.55 |
ENSDART00000058620
|
zp3d.1
|
zona pellucida glycoprotein 3d tandem duplicate 1 |
| chr2_-_50491234 | 3.55 |
ENSDART00000165678
|
mcm6l
|
MCM6 minichromosome maintenance deficient 6, like |
| chr7_-_47978449 | 3.54 |
ENSDART00000127007
ENSDART00000024062 |
cpeb1b
|
cytoplasmic polyadenylation element binding protein 1b |
| chr2_+_28199458 | 3.52 |
ENSDART00000150330
|
buc
|
bucky ball |
| chr10_+_16914003 | 3.51 |
ENSDART00000177906
|
UNC13B
|
unc-13 homolog B |
| chr7_+_67475765 | 3.43 |
ENSDART00000160086
|
zgc:162592
|
zgc:162592 |
| chr8_+_45326435 | 3.37 |
ENSDART00000134161
|
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr7_+_56787468 | 3.34 |
ENSDART00000024076
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr1_-_44892852 | 3.09 |
ENSDART00000160961
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr11_-_6442588 | 2.86 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr19_+_33552393 | 2.78 |
ENSDART00000043039
|
fam84b
|
family with sequence similarity 84, member B |
| chr1_-_44892600 | 2.76 |
ENSDART00000149155
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr5_-_23192997 | 2.70 |
ENSDART00000167629
|
rnf128a
|
ring finger protein 128a |
| chr7_+_58449163 | 2.70 |
ENSDART00000167166
|
zgc:56231
|
zgc:56231 |
| chr16_-_47446494 | 2.58 |
ENSDART00000032188
|
si:dkey-256h2.1
|
si:dkey-256h2.1 |
| chr21_-_34227158 | 2.56 |
ENSDART00000169218
ENSDART00000064320 ENSDART00000172381 |
alg13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr12_+_13704712 | 2.53 |
ENSDART00000152257
|
ppp1r16a
|
protein phosphatase 1, regulatory subunit 16A |
| chr5_-_28931727 | 2.53 |
ENSDART00000174697
|
arrdc1a
|
arrestin domain containing 1a |
| chr1_-_44893257 | 2.52 |
ENSDART00000160961
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr6_-_8125341 | 2.32 |
ENSDART00000139161
ENSDART00000140021 |
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr5_-_14993912 | 2.30 |
ENSDART00000085943
|
taok3a
|
TAO kinase 3a |
| chr23_-_18131502 | 2.26 |
ENSDART00000173075
|
zgc:92287
|
zgc:92287 |
| chr7_+_58449127 | 2.26 |
ENSDART00000167166
|
zgc:56231
|
zgc:56231 |
| chr6_+_135002 | 2.26 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr1_+_41762057 | 2.24 |
ENSDART00000137609
|
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr2_-_54224744 | 2.20 |
|
|
|
| chr14_-_32937536 | 2.14 |
ENSDART00000132850
|
stard14
|
START domain containing 14 |
| chr5_-_23192934 | 2.14 |
ENSDART00000021462
|
rnf128a
|
ring finger protein 128a |
| chr12_+_30389706 | 2.13 |
|
|
|
| chr14_-_7102535 | 2.09 |
ENSDART00000036463
|
dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr2_-_9854212 | 2.06 |
ENSDART00000112995
|
wu:fi34b01
|
wu:fi34b01 |
| chr1_-_52833379 | 2.05 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr10_+_21477579 | 2.03 |
ENSDART00000142447
|
etf1b
|
eukaryotic translation termination factor 1b |
| chr14_+_24543399 | 2.03 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr13_-_21541531 | 2.03 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr8_-_14516806 | 1.96 |
ENSDART00000057645
|
qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr5_+_58009274 | 1.94 |
ENSDART00000019561
|
zgc:171734
|
zgc:171734 |
| chr15_+_38397715 | 1.90 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr24_-_33894062 | 1.85 |
ENSDART00000079210
|
cdk5
|
cyclin-dependent kinase 5 |
| chr24_-_10253851 | 1.82 |
ENSDART00000127568
ENSDART00000106260 |
ankha
|
ANKH inorganic pyrophosphate transport regulator a |
| chr22_-_26537845 | 1.80 |
|
|
|
| chr17_-_31387062 | 1.78 |
|
|
|
| chr17_-_25313024 | 1.75 |
ENSDART00000082324
|
zpcx
|
zona pellucida protein C |
| chr5_-_32433490 | 1.74 |
|
|
|
| chr20_-_43846604 | 1.73 |
ENSDART00000150078
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr18_-_5167807 | 1.71 |
|
|
|
| chr3_-_20944579 | 1.70 |
ENSDART00000153739
|
nlk1
|
nemo-like kinase, type 1 |
| chr16_+_19831573 | 1.69 |
ENSDART00000135359
|
macc1
|
metastasis associated in colon cancer 1 |
| chr5_-_31738565 | 1.69 |
ENSDART00000017956
|
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr10_-_1933874 | 1.69 |
ENSDART00000101023
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr15_-_29393378 | 1.69 |
ENSDART00000114492
|
si:dkey-52l18.4
|
si:dkey-52l18.4 |
| chr22_-_10861268 | 1.64 |
ENSDART00000145229
|
arhgef18b
|
rho/rac guanine nucleotide exchange factor (GEF) 18b |
| chr2_+_9854294 | 1.64 |
|
|
|
| chr25_-_9889107 | 1.63 |
ENSDART00000137407
|
AL929493.1
|
ENSDARG00000093575 |
| chr10_-_31861975 | 1.62 |
ENSDART00000077785
|
vps26bl
|
vacuolar protein sorting 26 homolog B, like |
| chr6_+_47847753 | 1.60 |
ENSDART00000064842
|
padi2
|
peptidyl arginine deiminase, type II |
| chr24_+_14792755 | 1.57 |
ENSDART00000091735
|
dok6
|
docking protein 6 |
| chr3_+_42217236 | 1.54 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr10_+_37229202 | 1.53 |
ENSDART00000136510
|
ksr1a
|
kinase suppressor of ras 1a |
| chr12_-_30244042 | 1.53 |
ENSDART00000152878
|
tdrd1
|
tudor domain containing 1 |
| chr7_+_58448909 | 1.53 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr19_+_33551956 | 1.53 |
ENSDART00000043039
|
fam84b
|
family with sequence similarity 84, member B |
| chr23_-_31528548 | 1.51 |
|
|
|
| chr2_+_19588034 | 1.51 |
ENSDART00000163875
|
cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr9_-_38213786 | 1.50 |
|
|
|
| chr15_+_38397897 | 1.47 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr19_+_2942485 | 1.47 |
ENSDART00000177848
|
CABZ01066434.1
|
ENSDARG00000107451 |
| chr20_-_13729816 | 1.46 |
ENSDART00000078893
|
ezrb
|
ezrin b |
| chr19_+_7717962 | 1.46 |
ENSDART00000112404
|
cgnb
|
cingulin b |
| chr20_-_46079578 | 1.44 |
ENSDART00000153228
|
AL929237.3
|
ENSDARG00000096676 |
| chr17_-_36913213 | 1.42 |
ENSDART00000154981
|
senp6b
|
SUMO1/sentrin specific peptidase 6b |
| chr5_+_61389881 | 1.40 |
ENSDART00000141574
|
si:dkey-35m8.1
|
si:dkey-35m8.1 |
| chr7_+_19300487 | 1.39 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr2_-_17443642 | 1.38 |
ENSDART00000136207
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr9_+_38832959 | 1.37 |
ENSDART00000110651
|
slc12a8
|
solute carrier family 12, member 8 |
| chr1_+_46640008 | 1.34 |
ENSDART00000053284
|
bcl9
|
B-cell CLL/lymphoma 9 |
| chr1_-_44892558 | 1.34 |
ENSDART00000149155
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr10_-_1933761 | 1.33 |
ENSDART00000101023
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr2_+_1645259 | 1.33 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr1_+_46639773 | 1.33 |
|
|
|
| chr11_+_24076334 | 1.31 |
ENSDART00000017599
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr2_+_10858480 | 1.30 |
ENSDART00000091570
|
fam69aa
|
family with sequence similarity 69, member Aa |
| chr2_-_31817448 | 1.28 |
ENSDART00000170880
|
retreg1
|
reticulophagy regulator 1 |
| chr9_-_29128379 | 1.28 |
ENSDART00000101269
|
epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr4_-_12979951 | 1.25 |
ENSDART00000013839
|
tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr18_+_45669615 | 1.24 |
ENSDART00000150973
|
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr16_-_25317921 | 1.22 |
|
|
|
| chr2_-_31817409 | 1.21 |
ENSDART00000112763
|
retreg1
|
reticulophagy regulator 1 |
| chr6_-_12665211 | 1.21 |
ENSDART00000150887
|
ical1
|
islet cell autoantigen 1-like |
| chr14_+_20045326 | 1.20 |
ENSDART00000167637
|
aff2
|
AF4/FMR2 family, member 2 |
| chr10_-_31862004 | 1.18 |
ENSDART00000077785
|
vps26bl
|
vacuolar protein sorting 26 homolog B, like |
| chr1_+_47323244 | 1.16 |
|
|
|
| chr19_-_10295398 | 1.16 |
ENSDART00000148225
|
znf865
|
zinc finger protein 865 |
| chr4_-_12979905 | 1.15 |
ENSDART00000150674
|
tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr23_+_26153043 | 1.13 |
ENSDART00000054013
|
atp6ap1b
|
ATPase, H+ transporting, lysosomal accessory protein 1b |
| chr20_-_46079529 | 1.12 |
ENSDART00000153228
|
AL929237.3
|
ENSDARG00000096676 |
| chr5_-_56953587 | 1.12 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr9_-_5502510 | 1.11 |
ENSDART00000082260
|
abhd13
|
abhydrolase domain containing 13 |
| chr23_+_39961676 | 1.11 |
ENSDART00000161881
|
ENSDARG00000104435
|
ENSDARG00000104435 |
| chr2_+_9762781 | 1.11 |
ENSDART00000003465
|
gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr15_-_5636737 | 1.09 |
ENSDART00000114410
|
wdr62
|
WD repeat domain 62 |
| chr1_+_46639805 | 1.09 |
|
|
|
| chr5_+_32433624 | 1.09 |
|
|
|
| chr10_+_22065599 | 1.09 |
ENSDART00000143461
|
npm1a
|
nucleophosmin 1a (nucleolar phosphoprotein B23, numatrin) |
| chr5_+_3683209 | 1.08 |
ENSDART00000132056
|
ggnbp2
|
gametogenetin binding protein 2 |
| chr20_-_43846553 | 1.08 |
ENSDART00000150078
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr5_+_61711614 | 1.07 |
ENSDART00000082965
|
ABR
|
active BCR-related |
| chr12_+_33383407 | 1.04 |
ENSDART00000000069
|
slc9a3r1a
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1a |
| chr16_-_22394669 | 1.04 |
|
|
|
| chr2_+_52225622 | 1.03 |
ENSDART00000170353
|
ENSDARG00000103108
|
ENSDARG00000103108 |
| chr11_-_18078147 | 1.03 |
ENSDART00000177625
|
zgc:173545
|
zgc:173545 |
| chr3_-_26073798 | 1.03 |
ENSDART00000169123
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr18_+_27769446 | 1.02 |
|
|
|
| chr6_+_135255 | 1.01 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr4_+_13902603 | 1.00 |
ENSDART00000137549
|
pphln1
|
periphilin 1 |
| chr12_-_4264610 | 1.00 |
ENSDART00000152377
|
ca15b
|
carbonic anhydrase XVb |
| chr10_-_8839096 | 0.99 |
ENSDART00000080763
|
si:dkey-27b3.2
|
si:dkey-27b3.2 |
| chr8_-_22252628 | 0.99 |
ENSDART00000121513
|
nphp4
|
nephronophthisis 4 |
| chr15_-_29393320 | 0.98 |
ENSDART00000114492
|
si:dkey-52l18.4
|
si:dkey-52l18.4 |
| chr4_-_13615927 | 0.98 |
ENSDART00000138366
|
irf5
|
interferon regulatory factor 5 |
| chr19_-_32313943 | 0.97 |
ENSDART00000113797
|
zbtb10
|
zinc finger and BTB domain containing 10 |
| chr9_+_24310158 | 0.97 |
ENSDART00000101565
|
MAIP1
|
matrix AAA peptidase interacting protein 1 |
| chr2_+_41909094 | 0.97 |
ENSDART00000179428
|
CABZ01014197.2
|
ENSDARG00000107429 |
| chr15_+_35105420 | 0.94 |
ENSDART00000153840
|
si:ch73-95l15.5
|
si:ch73-95l15.5 |
| chr20_+_18803841 | 0.94 |
ENSDART00000152342
|
eif5
|
eukaryotic translation initiation factor 5 |
| chr4_+_4825409 | 0.94 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr13_+_31586024 | 0.93 |
ENSDART00000034745
|
prkcha
|
protein kinase C, eta, a |
| chr14_-_35932521 | 0.93 |
ENSDART00000158722
|
BX511223.1
|
ENSDARG00000101064 |
| chr8_-_49217824 | 0.93 |
|
|
|
| chr3_-_33362324 | 0.91 |
ENSDART00000020342
|
sgsm3
|
small G protein signaling modulator 3 |
| chr25_+_11212787 | 0.90 |
ENSDART00000159583
|
FQ312013.1
|
ENSDARG00000099473 |
| chr22_-_337759 | 0.89 |
|
|
|
| chr12_+_27613979 | 0.89 |
|
|
|
| chr2_+_43693486 | 0.88 |
|
|
|
| chr5_-_64432697 | 0.88 |
ENSDART00000165556
|
tor2a
|
torsin family 2, member A |
| chr5_-_56953716 | 0.88 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr11_-_11353309 | 0.87 |
ENSDART00000016677
|
zgc:77929
|
zgc:77929 |
| chr6_-_8125287 | 0.86 |
ENSDART00000139161
ENSDART00000140021 |
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr6_-_12786937 | 0.86 |
ENSDART00000157139
|
tmbim1a
|
transmembrane BAX inhibitor motif containing 1a |
| chr1_-_44892979 | 0.86 |
ENSDART00000160961
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr8_-_17736487 | 0.85 |
ENSDART00000063592
ENSDART00000175064 |
prkcz
|
protein kinase C, zeta |
| chr24_-_12622124 | 0.84 |
|
|
|
| chr21_-_16979722 | 0.84 |
|
|
|
| chr3_-_26052785 | 0.84 |
ENSDART00000147517
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr3_+_42217187 | 0.83 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr2_+_43355846 | 0.83 |
ENSDART00000056402
|
pard3ab
|
par-3 family cell polarity regulator alpha, b |
| chr23_+_37124543 | 0.83 |
ENSDART00000155706
|
CR762407.2
|
ENSDARG00000097111 |
| chr19_-_32314264 | 0.83 |
|
|
|
| chr1_-_52833426 | 0.82 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr19_+_33392145 | 0.82 |
ENSDART00000171782
|
spire1a
|
spire-type actin nucleation factor 1a |
| chr2_+_9762627 | 0.81 |
ENSDART00000003465
|
gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr9_-_30174641 | 0.80 |
ENSDART00000134157
ENSDART00000089206 |
il1rapl1a
|
interleukin 1 receptor accessory protein-like 1a |
| chr22_-_4787016 | 0.80 |
ENSDART00000140313
|
tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr6_+_47847593 | 0.80 |
ENSDART00000064842
|
padi2
|
peptidyl arginine deiminase, type II |
| chr18_+_13280623 | 0.80 |
ENSDART00000080709
|
plcg2
|
phospholipase C, gamma 2 |
| chr1_+_47323043 | 0.80 |
|
|
|
| chr18_-_35861412 | 0.80 |
ENSDART00000088488
|
opa3
|
optic atrophy 3 |
| chr16_+_34157948 | 0.80 |
ENSDART00000140552
|
tcea3
|
transcription elongation factor A (SII), 3 |
| chr7_+_67475893 | 0.79 |
ENSDART00000160086
|
zgc:162592
|
zgc:162592 |
| chr22_-_10861329 | 0.75 |
ENSDART00000145229
|
arhgef18b
|
rho/rac guanine nucleotide exchange factor (GEF) 18b |
| chr11_+_24076371 | 0.75 |
ENSDART00000166045
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr1_-_44621977 | 0.74 |
|
|
|
| chr11_-_6442547 | 0.74 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr25_+_27300835 | 0.73 |
ENSDART00000103519
|
wasla
|
Wiskott-Aldrich syndrome-like a |
| chr9_-_45104857 | 0.73 |
ENSDART00000131252
|
zgc:66484
|
zgc:66484 |
| chr3_-_26052601 | 0.72 |
ENSDART00000147517
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr21_+_16979503 | 0.71 |
ENSDART00000065755
|
gpn3
|
GPN-loop GTPase 3 |
| chr18_+_22299937 | 0.70 |
|
|
|
| chr9_+_18565169 | 0.69 |
|
|
|
| chr18_+_13280400 | 0.69 |
ENSDART00000080709
|
plcg2
|
phospholipase C, gamma 2 |
| chr25_+_27301039 | 0.68 |
ENSDART00000149456
|
wasla
|
Wiskott-Aldrich syndrome-like a |
| chr6_+_134956 | 0.68 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr17_-_36913302 | 0.66 |
ENSDART00000154981
|
senp6b
|
SUMO1/sentrin specific peptidase 6b |
| chr11_-_6442490 | 0.65 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr23_+_39231080 | 0.65 |
|
|
|
| chr5_+_57254656 | 0.64 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr22_-_26231695 | 0.63 |
ENSDART00000142821
|
ccdc130
|
coiled-coil domain containing 130 |
| chr14_+_21874825 | 0.63 |
ENSDART00000114750
|
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr18_-_35861482 | 0.63 |
ENSDART00000088488
|
opa3
|
optic atrophy 3 |
| chr16_-_25318045 | 0.63 |
ENSDART00000058943
|
ENSDARG00000040280
|
ENSDARG00000040280 |
| chr14_-_30747372 | 0.63 |
ENSDART00000002250
|
hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr23_-_19905270 | 0.62 |
ENSDART00000054479
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr7_-_50094750 | 0.62 |
ENSDART00000134941
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr21_-_34227100 | 0.62 |
ENSDART00000124649
|
alg13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr5_-_21548930 | 0.62 |
ENSDART00000002938
|
mtmr8
|
myotubularin related protein 8 |
| chr8_-_14516930 | 0.62 |
ENSDART00000057645
|
qsox1
|
quiescin Q6 sulfhydryl oxidase 1 |
| chr3_-_33362384 | 0.61 |
ENSDART00000020342
|
sgsm3
|
small G protein signaling modulator 3 |
| chr20_-_25731057 | 0.61 |
|
|
|
| chr21_-_16980069 | 0.60 |
|
|
|
| chr6_-_10492724 | 0.60 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr12_+_13704914 | 0.60 |
ENSDART00000152257
|
ppp1r16a
|
protein phosphatase 1, regulatory subunit 16A |
| chr8_+_44619220 | 0.59 |
ENSDART00000063392
|
lsm1
|
LSM1, U6 small nuclear RNA associated |
| chr11_-_27378184 | 0.59 |
ENSDART00000157337
|
CR931782.1
|
ENSDARG00000097455 |
| chr8_+_45326523 | 0.59 |
ENSDART00000145011
|
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 1.0 | 3.0 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.9 | 3.5 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.9 | 10.6 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.6 | 2.4 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.6 | 2.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.6 | 1.7 | GO:1905207 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.6 | 2.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.5 | 2.5 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.5 | 3.2 | GO:0045453 | bone resorption(GO:0045453) |
| 0.4 | 2.1 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.4 | 2.1 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.4 | 4.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.4 | 3.2 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.3 | 2.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.3 | 1.0 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.3 | 1.3 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.3 | 1.3 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.3 | 5.3 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.3 | 3.5 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) mitotic DNA replication(GO:1902969) |
| 0.3 | 1.1 | GO:0051029 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.2 | 3.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 | 4.7 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.2 | 1.3 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.2 | 1.3 | GO:0021744 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.2 | 2.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.2 | 0.8 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.2 | 1.0 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.2 | 2.2 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.2 | 0.7 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 1.5 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 1.3 | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) |
| 0.1 | 1.0 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.1 | 0.8 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.1 | 4.4 | GO:0048599 | oocyte development(GO:0048599) |
| 0.1 | 1.4 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 3.6 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 1.4 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.9 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 1.8 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 1.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.4 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 2.5 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.1 | 2.4 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.9 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 13.4 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.1 | 0.1 | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) |
| 0.1 | 0.6 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.6 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.5 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 4.4 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.1 | 0.2 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 3.1 | GO:0035304 | regulation of protein dephosphorylation(GO:0035304) |
| 0.1 | 2.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.4 | GO:0050881 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.0 | 1.0 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.9 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.6 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 1.0 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.9 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.0 | 0.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 2.3 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 0.8 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.3 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.2 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.0 | 1.8 | GO:0045765 | regulation of angiogenesis(GO:0045765) |
| 0.0 | 0.1 | GO:0007063 | regulation of sister chromatid cohesion(GO:0007063) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 1.6 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.6 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 2.1 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.5 | 2.5 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.3 | 3.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 1.5 | GO:0071546 | pi-body(GO:0071546) |
| 0.2 | 2.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.2 | 3.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 1.3 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.2 | 3.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 10.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.2 | 0.8 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 2.1 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 4.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 1.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 1.0 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 0.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.0 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 2.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 3.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 4.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.6 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.2 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 1.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 3.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.6 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.4 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.9 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.1 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.5 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.2 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) alpha1-adrenergic receptor activity(GO:0004937) |
| 0.9 | 3.5 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.8 | 2.5 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.6 | 2.6 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.6 | 1.8 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.6 | 2.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.5 | 2.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.5 | 2.5 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.4 | 2.6 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.4 | 3.2 | GO:0003993 | acid phosphatase activity(GO:0003993) ferric iron binding(GO:0008199) |
| 0.4 | 3.5 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.4 | 1.5 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.4 | 4.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.3 | 2.4 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.3 | 1.0 | GO:0031730 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) CCR5 chemokine receptor binding(GO:0031730) |
| 0.3 | 5.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.3 | 4.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.3 | 3.0 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.3 | 0.8 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.2 | 4.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 0.7 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.2 | 1.4 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.2 | 10.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 0.7 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.2 | 0.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 1.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 2.3 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 1.4 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 2.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 2.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.5 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 3.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 3.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 2.4 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 2.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.3 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.1 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.9 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.6 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 1.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.5 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 2.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.6 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 4.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 2.5 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.0 | 0.5 | GO:0004620 | phospholipase activity(GO:0004620) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 1.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 1.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 1.9 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 2.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 1.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 1.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.3 | 2.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 1.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 1.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 3.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 0.6 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 3.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 1.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 0.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |