Project
DANIO-CODE
Navigation
Downloads
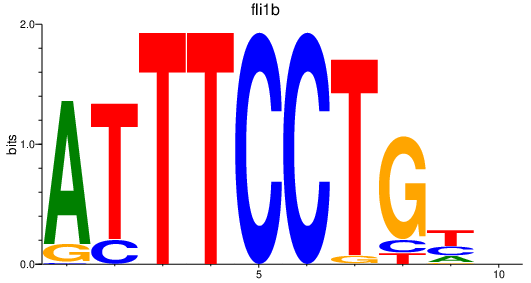
Results for fli1b
Z-value: 0.95
Transcription factors associated with fli1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
fli1b
|
ENSDARG00000040080 | Fli-1 proto-oncogene, ETS transcription factor b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| fli1b | dr10_dc_chr16_+_42067930_42068014 | 0.91 | 1.3e-06 | Click! |
Activity profile of fli1b motif
Sorted Z-values of fli1b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of fli1b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_9924987 | 3.45 |
ENSDART00000005015
|
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr5_-_62747812 | 3.42 |
|
|
|
| chr7_-_25422583 | 2.61 |
ENSDART00000135415
|
dysf
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) |
| chr3_+_31542311 | 2.39 |
|
|
|
| chr8_-_7049946 | 2.38 |
ENSDART00000045669
ENSDART00000162950 |
KRT73
|
keratin 73 |
| chr7_+_19765237 | 2.27 |
ENSDART00000100808
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr5_-_1325831 | 2.22 |
|
|
|
| chr18_+_38307946 | 2.17 |
ENSDART00000134247
|
lmo2
|
LIM domain only 2 (rhombotin-like 1) |
| chr11_+_8119829 | 2.14 |
ENSDART00000011183
|
prkacba
|
protein kinase, cAMP-dependent, catalytic, beta a |
| chr5_-_62747914 | 2.10 |
|
|
|
| chr15_-_4537178 | 1.89 |
ENSDART00000155619
ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr23_+_13911019 | 1.78 |
ENSDART00000168545
|
zbtb46
|
zinc finger and BTB domain containing 46 |
| chr9_-_34129128 | 1.75 |
|
|
|
| chr5_-_67043355 | 1.75 |
ENSDART00000134386
|
arhgap31
|
Rho GTPase activating protein 31 |
| chr5_-_34016383 | 1.71 |
ENSDART00000050271
ENSDART00000097975 |
hexb
|
hexosaminidase B (beta polypeptide) |
| chr18_-_25002972 | 1.70 |
ENSDART00000163449
|
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr14_+_49921861 | 1.67 |
ENSDART00000173240
|
zgc:154054
|
zgc:154054 |
| chr19_-_2399429 | 1.64 |
ENSDART00000043595
|
twist1a
|
twist family bHLH transcription factor 1a |
| chr22_+_5657904 | 1.64 |
ENSDART00000138102
|
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr23_+_36822250 | 1.59 |
|
|
|
| chr19_-_325337 | 1.58 |
ENSDART00000147635
|
gpd1c
|
glycerol-3-phosphate dehydrogenase 1c |
| chr16_-_39620777 | 1.57 |
ENSDART00000039832
|
tgfbr2b
|
transforming growth factor beta receptor 2b |
| chr14_+_13147115 | 1.56 |
ENSDART00000054849
|
pls3
|
plastin 3 (T isoform) |
| chr5_+_36487425 | 1.56 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr4_-_17735989 | 1.47 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr5_-_41672394 | 1.47 |
ENSDART00000164363
|
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr5_-_15774816 | 1.46 |
ENSDART00000090684
|
kremen1
|
kringle containing transmembrane protein 1 |
| chr4_-_22590638 | 1.45 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr5_+_4033390 | 1.40 |
ENSDART00000149185
|
sat1a.2
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 2 |
| chr21_-_11539798 | 1.39 |
ENSDART00000144770
|
cast
|
calpastatin |
| chr20_+_20600211 | 1.37 |
ENSDART00000036124
|
six1b
|
SIX homeobox 1b |
| chr9_+_54692834 | 1.36 |
|
|
|
| chr7_+_34482282 | 1.36 |
|
|
|
| chr22_-_13139894 | 1.35 |
ENSDART00000105762
|
ahr2
|
aryl hydrocarbon receptor 2 |
| chr19_-_35512932 | 1.35 |
|
|
|
| chr4_+_3344998 | 1.34 |
ENSDART00000075320
|
nampta
|
nicotinamide phosphoribosyltransferase a |
| chr22_-_23641813 | 1.31 |
ENSDART00000159622
|
cfh
|
complement factor H |
| chr1_+_49041978 | 1.31 |
ENSDART00000140824
|
si:ch211-149l1.2
|
si:ch211-149l1.2 |
| chr17_-_29102320 | 1.30 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr15_-_2689005 | 1.30 |
ENSDART00000063325
|
cldnf
|
claudin f |
| chr13_-_24181106 | 1.30 |
ENSDART00000004420
|
rab4a
|
RAB4a, member RAS oncogene family |
| chr16_+_1338291 | 1.28 |
ENSDART00000149299
|
cers2b
|
ceramide synthase 2b |
| chr7_+_22552724 | 1.27 |
ENSDART00000101459
ENSDART00000159743 |
pygmb
|
phosphorylase, glycogen, muscle b |
| chr7_+_67244332 | 1.26 |
ENSDART00000170322
|
rpl13
|
ribosomal protein L13 |
| chr23_+_17939650 | 1.25 |
ENSDART00000162822
|
naca
|
nascent polypeptide-associated complex alpha subunit |
| chr24_+_36746202 | 1.25 |
ENSDART00000159017
|
si:ch73-334d15.4
|
si:ch73-334d15.4 |
| chr8_-_40231417 | 1.24 |
ENSDART00000162020
|
kdm2ba
|
lysine (K)-specific demethylase 2Ba |
| chr24_+_5906087 | 1.24 |
ENSDART00000131768
|
abi1a
|
abl-interactor 1a |
| chr18_+_23004984 | 1.23 |
ENSDART00000171871
|
cbfb
|
core-binding factor, beta subunit |
| chr2_+_22839514 | 1.23 |
ENSDART00000167915
|
lrrc8da
|
leucine rich repeat containing 8 family, member Da |
| chr19_-_35739239 | 1.21 |
|
|
|
| chr11_-_25803101 | 1.21 |
ENSDART00000088888
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr3_+_13043951 | 1.21 |
|
|
|
| chr19_-_35513216 | 1.21 |
|
|
|
| chr16_+_42067930 | 1.20 |
ENSDART00000102789
|
fli1b
|
Fli-1 proto-oncogene, ETS transcription factor b |
| chr22_-_120677 | 1.18 |
|
|
|
| chr13_+_11913290 | 1.18 |
ENSDART00000079398
|
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr24_-_10928910 | 1.17 |
ENSDART00000127398
|
CR753886.1
|
ENSDARG00000090548 |
| chr10_+_9764591 | 1.16 |
ENSDART00000091780
|
rc3h2
|
ring finger and CCCH-type domains 2 |
| chr25_+_7660590 | 1.15 |
ENSDART00000155016
|
dgkzb
|
diacylglycerol kinase, zeta b |
| chr20_-_36897042 | 1.15 |
ENSDART00000015190
|
ptrhd1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr12_+_27370834 | 1.14 |
ENSDART00000105661
|
meox1
|
mesenchyme homeobox 1 |
| chr17_+_1610578 | 1.14 |
ENSDART00000082101
|
ppp2r5ca
|
protein phosphatase 2, regulatory subunit B', gamma a |
| chr2_-_3317258 | 1.14 |
ENSDART00000167944
|
wnt3a
|
wingless-type MMTV integration site family, member 3A |
| chr21_+_30434147 | 1.13 |
ENSDART00000147375
|
snx12
|
sorting nexin 12 |
| chr7_-_64637419 | 1.11 |
ENSDART00000020456
|
mmp15b
|
matrix metallopeptidase 15b |
| chr6_+_30681170 | 1.09 |
ENSDART00000112294
|
ttc22
|
tetratricopeptide repeat domain 22 |
| chr9_-_23346038 | 1.08 |
ENSDART00000135461
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr12_-_28840796 | 1.07 |
|
|
|
| chr13_+_23065500 | 1.06 |
ENSDART00000158370
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr23_-_15807415 | 1.06 |
ENSDART00000178557
|
BX005011.3
|
ENSDARG00000108083 |
| chr5_+_18740715 | 1.06 |
ENSDART00000089173
|
atp8b5a
|
ATPase, class I, type 8B, member 5a |
| chr9_-_31714290 | 1.06 |
ENSDART00000127214
|
tmtc4
|
transmembrane and tetratricopeptide repeat containing 4 |
| chr18_+_91971 | 1.05 |
|
|
|
| chr13_+_16130212 | 1.05 |
ENSDART00000125813
|
mocos
|
molybdenum cofactor sulfurase |
| chr14_-_31128891 | 1.05 |
|
|
|
| chr13_-_39365252 | 1.05 |
ENSDART00000019379
|
marveld1
|
MARVEL domain containing 1 |
| chr16_+_38327295 | 1.05 |
ENSDART00000085143
|
bnipl
|
BCL2/adenovirus E1B 19kD interacting protein, like |
| chr16_-_36991800 | 1.04 |
ENSDART00000171351
|
aldh5a1
|
aldehyde dehydrogenase 5 family, member A1 (succinate-semialdehyde dehydrogenase) |
| KN149998v1_+_54953 | 1.04 |
|
|
|
| chr23_+_13910925 | 1.04 |
ENSDART00000168545
|
zbtb46
|
zinc finger and BTB domain containing 46 |
| chr16_-_26803204 | 1.03 |
ENSDART00000162665
|
esrp1
|
epithelial splicing regulatory protein 1 |
| chr19_+_33267939 | 1.03 |
ENSDART00000176726
|
rpl30
|
ribosomal protein L30 |
| chr16_-_42063851 | 1.02 |
ENSDART00000045403
|
etv2
|
ets variant 2 |
| chr13_+_25298383 | 1.01 |
|
|
|
| chr21_-_35806638 | 1.01 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr3_+_24327586 | 1.00 |
ENSDART00000153551
|
cbx6b
|
chromobox homolog 6b |
| chr9_-_7695437 | 1.00 |
ENSDART00000102715
|
tuba8l3
|
tubulin, alpha 8 like 3 |
| chr25_+_18460166 | 1.00 |
ENSDART00000073726
|
cav2
|
caveolin 2 |
| chr10_+_22759607 | 1.00 |
|
|
|
| chr20_+_16843502 | 1.00 |
ENSDART00000050308
|
calm1b
|
calmodulin 1b |
| chr17_+_3842688 | 0.99 |
ENSDART00000170822
|
tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr7_+_73408688 | 0.99 |
ENSDART00000159745
|
PCP4L1
|
Purkinje cell protein 4 like 1 |
| chr19_+_34114412 | 0.99 |
ENSDART00000162517
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr14_-_48951428 | 0.99 |
ENSDART00000157785
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr11_+_26371444 | 0.99 |
ENSDART00000042322
|
map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr9_-_1949904 | 0.98 |
ENSDART00000082355
|
hoxd4a
|
homeobox D4a |
| chr24_+_9272045 | 0.97 |
ENSDART00000132724
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr24_+_42884 | 0.97 |
ENSDART00000122785
|
zgc:152808
|
zgc:152808 |
| chr13_+_11912981 | 0.97 |
ENSDART00000158244
|
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr24_-_6048914 | 0.97 |
ENSDART00000146830
ENSDART00000021981 |
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr8_+_54171992 | 0.96 |
ENSDART00000122692
|
CABZ01112317.1
|
ENSDARG00000086057 |
| chr14_+_26498814 | 0.96 |
ENSDART00000137695
|
klhl4
|
kelch-like family member 4 |
| chr25_-_20283460 | 0.95 |
ENSDART00000067454
|
kctd15a
|
potassium channel tetramerization domain containing 15a |
| chr6_+_1895734 | 0.95 |
ENSDART00000109679
|
quo
|
quattro |
| chr21_+_25729090 | 0.94 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr9_-_9693163 | 0.94 |
ENSDART00000018228
|
gsk3b
|
glycogen synthase kinase 3 beta |
| chr22_-_14090538 | 0.94 |
ENSDART00000105717
|
aox5
|
aldehyde oxidase 5 |
| chr14_-_425659 | 0.93 |
|
|
|
| chr7_-_35042805 | 0.93 |
ENSDART00000005053
|
slc12a4
|
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr5_+_31590746 | 0.93 |
|
|
|
| chr23_-_29138952 | 0.93 |
ENSDART00000002812
|
casz1
|
castor zinc finger 1 |
| chr16_+_53238110 | 0.92 |
ENSDART00000102170
|
CABZ01053976.1
|
ENSDARG00000069929 |
| chr7_+_13750329 | 0.92 |
ENSDART00000091470
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr5_-_31275147 | 0.91 |
ENSDART00000098160
|
tmem119b
|
transmembrane protein 119b |
| chr18_+_48426375 | 0.90 |
|
|
|
| chr20_-_40217185 | 0.89 |
|
|
|
| chr6_-_12224562 | 0.89 |
ENSDART00000090266
ENSDART00000144028 |
gpd2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr17_+_381715 | 0.89 |
ENSDART00000162898
|
si:rp71-62i8.1
|
si:rp71-62i8.1 |
| chr3_+_23638277 | 0.89 |
ENSDART00000110682
|
hoxb1a
|
homeobox B1a |
| chr11_+_43924640 | 0.88 |
ENSDART00000179206
|
gnb4b
|
guanine nucleotide binding protein (G protein), beta polypeptide 4b |
| chr6_+_19795100 | 0.87 |
|
|
|
| chr3_+_22246697 | 0.87 |
ENSDART00000155597
|
arhgap27l
|
Rho GTPase activating protein 27, like |
| chr5_-_58269742 | 0.86 |
ENSDART00000122413
|
mcama
|
melanoma cell adhesion molecule a |
| chr12_+_36716775 | 0.86 |
ENSDART00000048927
|
cox10
|
COX10 heme A:farnesyltransferase cytochrome c oxidase assembly factor |
| chr19_-_7069920 | 0.86 |
ENSDART00000145741
|
znf384l
|
zinc finger protein 384 like |
| chr16_-_16582360 | 0.86 |
|
|
|
| chr23_-_24755654 | 0.86 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr14_-_32553399 | 0.85 |
ENSDART00000169380
|
slc25a43
|
solute carrier family 25, member 43 |
| chr21_+_25034997 | 0.85 |
ENSDART00000167523
|
dixdc1b
|
DIX domain containing 1b |
| chr17_-_39237265 | 0.85 |
ENSDART00000050534
|
crim1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
| chr5_+_30143469 | 0.85 |
ENSDART00000138292
|
ftr83
|
finTRIM family, member 83 |
| chr1_+_35933511 | 0.85 |
ENSDART00000010632
|
ednraa
|
endothelin receptor type Aa |
| chr24_+_18804086 | 0.85 |
ENSDART00000106186
|
prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr25_+_34069931 | 0.84 |
|
|
|
| chr10_-_35036872 | 0.84 |
ENSDART00000170625
|
smad9
|
SMAD family member 9 |
| chr1_-_51862897 | 0.83 |
ENSDART00000136469
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr5_-_43686611 | 0.82 |
ENSDART00000146080
|
si:ch73-337l15.2
|
si:ch73-337l15.2 |
| chr3_-_23558171 | 0.82 |
|
|
|
| chr20_+_20287015 | 0.82 |
ENSDART00000002507
|
rhoj
|
ras homolog family member J |
| chr5_+_68943914 | 0.82 |
ENSDART00000159594
|
mthfd2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr1_-_18695214 | 0.82 |
|
|
|
| chr3_+_23540206 | 0.82 |
|
|
|
| chr11_+_5929544 | 0.81 |
ENSDART00000104364
|
rps15
|
ribosomal protein S15 |
| chr3_-_50386208 | 0.81 |
ENSDART00000156709
|
BX088529.1
|
ENSDARG00000097178 |
| chr4_+_7668939 | 0.81 |
ENSDART00000149218
|
elk3
|
ELK3, ETS-domain protein |
| chr17_+_11264847 | 0.81 |
ENSDART00000153602
|
CU076099.1
|
ENSDARG00000097666 |
| chr8_-_19250414 | 0.81 |
ENSDART00000100473
|
ENSDARG00000013711
|
ENSDARG00000013711 |
| chr20_-_20502615 | 0.80 |
|
|
|
| chr9_-_12726136 | 0.80 |
|
|
|
| chr5_-_54954988 | 0.79 |
ENSDART00000142912
|
hnrpkl
|
heterogeneous nuclear ribonucleoprotein K, like |
| chr23_+_31181140 | 0.79 |
ENSDART00000103448
|
tbx18
|
T-box 18 |
| chr7_-_64637051 | 0.79 |
ENSDART00000020456
|
mmp15b
|
matrix metallopeptidase 15b |
| chr14_-_33936261 | 0.78 |
ENSDART00000021437
|
gria1a
|
glutamate receptor, ionotropic, AMPA 1a |
| chr3_+_25933164 | 0.78 |
ENSDART00000143697
|
si:dkeyp-69e1.8
|
si:dkeyp-69e1.8 |
| chr17_+_31346338 | 0.77 |
ENSDART00000062900
|
itpka
|
inositol-trisphosphate 3-kinase A |
| chr23_-_28286971 | 0.77 |
|
|
|
| chr7_-_38418301 | 0.77 |
ENSDART00000167209
|
aplnr2
|
apelin receptor 2 |
| chr1_-_7456961 | 0.75 |
ENSDART00000152295
|
FAM83G
|
family with sequence similarity 83 member G |
| chr1_-_46052304 | 0.75 |
ENSDART00000142880
|
BX294379.1
|
ENSDARG00000092950 |
| chr13_+_28689749 | 0.75 |
ENSDART00000101653
|
CU639469.1
|
ENSDARG00000062790 |
| chr21_+_43204049 | 0.74 |
ENSDART00000151748
|
aff4
|
AF4/FMR2 family, member 4 |
| chr20_+_26408687 | 0.74 |
|
|
|
| chr5_+_45677439 | 0.74 |
ENSDART00000045598
|
zgc:110626
|
zgc:110626 |
| chr5_-_62816208 | 0.74 |
ENSDART00000097325
|
c5
|
complement component 5 |
| chr20_-_475417 | 0.73 |
ENSDART00000032212
|
fynrk
|
fyn-related Src family tyrosine kinase |
| KN149859v1_+_16751 | 0.73 |
ENSDART00000167610
|
ENSDARG00000103399
|
ENSDARG00000103399 |
| chr1_+_25928481 | 0.73 |
ENSDART00000158193
|
coro2a
|
coronin, actin binding protein, 2A |
| chr11_-_3968366 | 0.73 |
ENSDART00000171093
|
glt8d1
|
glycosyltransferase 8 domain containing 1 |
| chr8_+_36888422 | 0.73 |
|
|
|
| chr3_-_61104221 | 0.73 |
ENSDART00000155414
|
tecpr1b
|
tectonin beta-propeller repeat containing 1b |
| chr11_-_36088889 | 0.73 |
ENSDART00000146495
|
psma5
|
proteasome subunit alpha 5 |
| chr19_+_5375413 | 0.73 |
ENSDART00000141237
|
si:dkeyp-113d7.10
|
si:dkeyp-113d7.10 |
| chr9_+_36051713 | 0.72 |
ENSDART00000134447
|
rcan1a
|
regulator of calcineurin 1a |
| chr12_+_28739504 | 0.72 |
ENSDART00000152991
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr13_+_41998500 | 0.72 |
ENSDART00000074707
|
cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr23_+_19018062 | 0.72 |
ENSDART00000104487
|
cox4i2
|
cytochrome c oxidase subunit IV isoform 2 |
| chr9_-_56644466 | 0.71 |
ENSDART00000149851
|
rpl31
|
ribosomal protein L31 |
| chr10_+_16589111 | 0.71 |
ENSDART00000084278
|
isoc1
|
isochorismatase domain containing 1 |
| chr20_+_25441689 | 0.71 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr4_+_22757046 | 0.71 |
|
|
|
| chr10_+_4717800 | 0.71 |
ENSDART00000161789
|
palm2
|
paralemmin 2 |
| chr18_-_25659136 | 0.70 |
ENSDART00000138861
|
si:ch211-13k12.2
|
si:ch211-13k12.2 |
| chr5_-_67043168 | 0.70 |
ENSDART00000134386
|
arhgap31
|
Rho GTPase activating protein 31 |
| chr13_+_32013916 | 0.70 |
ENSDART00000020270
|
osr1
|
odd-skipped related transciption factor 1 |
| chr23_+_36007936 | 0.70 |
ENSDART00000128533
|
hoxc3a
|
homeo box C3a |
| chr5_-_12213712 | 0.70 |
ENSDART00000099749
|
CLDN22
|
claudin 22 |
| chr16_-_48672056 | 0.70 |
|
|
|
| chr10_-_11053655 | 0.69 |
ENSDART00000132995
|
ak3
|
adenylate kinase 3 |
| chr8_+_15201828 | 0.69 |
ENSDART00000132216
|
paox1
|
polyamine oxidase (exo-N4-amino) 1 |
| chr8_+_11604334 | 0.69 |
ENSDART00000004288
|
ift81
|
intraflagellar transport 81 homolog |
| chr8_-_12365342 | 0.68 |
ENSDART00000113286
|
phf19
|
PHD finger protein 19 |
| chr13_+_24271932 | 0.67 |
ENSDART00000043002
|
rab1ab
|
RAB1A, member RAS oncogene family b |
| chr10_+_4987494 | 0.67 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr14_+_24637864 | 0.67 |
ENSDART00000170871
|
ppargc1b
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr11_-_176933 | 0.66 |
ENSDART00000172920
|
cdk4
|
cyclin-dependent kinase 4 |
| chr5_-_31275259 | 0.66 |
ENSDART00000098160
|
tmem119b
|
transmembrane protein 119b |
| chr7_+_19122698 | 0.66 |
ENSDART00000162700
|
snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr10_+_7043530 | 0.66 |
|
|
|
| chr10_+_29351676 | 0.66 |
ENSDART00000088973
|
sytl2a
|
synaptotagmin-like 2a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0050765 | monocyte activation involved in immune response(GO:0002280) negative regulation of phagocytosis(GO:0050765) |
| 0.7 | 2.0 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.4 | 1.6 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.4 | 1.6 | GO:0022602 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.4 | 2.0 | GO:0048901 | anterior lateral line neuromast development(GO:0048901) anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.4 | 2.2 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.4 | 1.8 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.3 | 0.3 | GO:0042116 | macrophage activation involved in immune response(GO:0002281) macrophage activation(GO:0042116) |
| 0.3 | 1.4 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.3 | 1.2 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.3 | 0.9 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.3 | 0.9 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.3 | 1.0 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.3 | 1.0 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.2 | 0.9 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 0.8 | GO:0043586 | tongue development(GO:0043586) tongue morphogenesis(GO:0043587) taste bud development(GO:0061193) taste bud morphogenesis(GO:0061194) taste bud formation(GO:0061195) |
| 0.2 | 0.5 | GO:1900060 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.2 | 0.9 | GO:0021561 | facial nerve development(GO:0021561) |
| 0.2 | 1.1 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.2 | 1.0 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.2 | 0.7 | GO:0035776 | pronephric proximal tubule development(GO:0035776) |
| 0.2 | 0.3 | GO:0050764 | regulation of phagocytosis(GO:0050764) |
| 0.2 | 0.6 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.2 | 1.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 1.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 1.4 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 0.6 | GO:0043651 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 1.9 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 0.9 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.4 | GO:0014005 | microglia development(GO:0014005) |
| 0.1 | 1.6 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.9 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 1.5 | GO:0000272 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 1.7 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.1 | 0.5 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.1 | 0.4 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 0.6 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.7 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 2.1 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.1 | 0.4 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.1 | 1.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.2 | GO:0070528 | protein kinase C signaling(GO:0070528) |
| 0.1 | 0.3 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.1 | 0.4 | GO:0072425 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.1 | 0.3 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 0.8 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 0.4 | GO:0046888 | negative regulation of hormone secretion(GO:0046888) |
| 0.1 | 0.4 | GO:0033499 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 0.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 1.1 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.4 | GO:1901492 | regulation of lymphangiogenesis(GO:1901490) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 2.2 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.9 | GO:0042310 | vasoconstriction(GO:0042310) |
| 0.1 | 1.1 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 2.1 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.1 | 0.5 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.1 | 0.8 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 1.1 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.3 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 2.1 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.1 | 0.4 | GO:0072530 | purine-containing compound transmembrane transport(GO:0072530) |
| 0.1 | 0.5 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 1.0 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 1.3 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 0.3 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.2 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.7 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.2 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.1 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.2 | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response(GO:1900101) negative regulation of endoplasmic reticulum unfolded protein response(GO:1900102) regulation of IRE1-mediated unfolded protein response(GO:1903894) negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.1 | 2.6 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 0.5 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.1 | 0.4 | GO:0048041 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.1 | 0.4 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.7 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 0.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.2 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.5 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.2 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.0 | 0.6 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 1.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.6 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.8 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.8 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.7 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0070863 | intestinal cholesterol absorption(GO:0030299) regulation of protein exit from endoplasmic reticulum(GO:0070861) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.8 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 1.1 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.5 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 2.4 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 2.1 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.9 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 1.8 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.5 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 1.4 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.2 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.4 | GO:0060606 | neural tube closure(GO:0001843) tube closure(GO:0060606) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.6 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 1.4 | GO:0001817 | regulation of cytokine production(GO:0001817) |
| 0.0 | 1.2 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 1.8 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.5 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.7 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 1.2 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 0.4 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.3 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.4 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.0 | 0.3 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 1.1 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.1 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0044331 | cell-cell adhesion mediated by cadherin(GO:0044331) regulation of cell-cell adhesion mediated by cadherin(GO:2000047) |
| 0.0 | 0.2 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.2 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.3 | GO:0031623 | receptor internalization(GO:0031623) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.2 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 0.6 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 0.3 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.4 | GO:0030032 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 0.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 2.1 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.6 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.1 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 0.6 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.3 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.0 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.0 | 0.3 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.6 | GO:0016331 | morphogenesis of embryonic epithelium(GO:0016331) |
| 0.0 | 1.9 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 0.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.6 | GO:0007422 | peripheral nervous system development(GO:0007422) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.4 | 2.7 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.3 | 0.9 | GO:0098594 | mucin granule(GO:0098594) |
| 0.3 | 0.8 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.2 | 1.2 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.2 | 3.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 2.1 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.2 | 0.6 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 0.5 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 0.5 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.4 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.8 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 1.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.5 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.1 | 0.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 0.6 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.0 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 1.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.7 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.4 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 3.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 3.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 1.8 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 1.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 2.2 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.5 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 1.6 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 1.1 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 1.6 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.2 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.9 | GO:0098791 | Golgi subcompartment(GO:0098791) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 1.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 4.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.5 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.8 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0005026 | transforming growth factor beta receptor activity, type II(GO:0005026) type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.5 | 1.5 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.4 | 1.6 | GO:0019808 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.4 | 1.1 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.4 | 1.8 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.3 | 1.7 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.3 | 0.9 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.3 | 2.1 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 1.7 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.3 | 0.8 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.3 | 1.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.3 | 1.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.3 | 1.3 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.2 | 0.7 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.2 | 0.7 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.2 | 0.6 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.2 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 0.9 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.2 | 2.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 0.9 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 0.5 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 1.0 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.2 | 1.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 1.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.2 | 0.8 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.2 | 0.8 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.2 | 1.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.2 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 0.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 1.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.6 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.8 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 1.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.8 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 1.0 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.4 | GO:0032028 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.1 | 0.3 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 1.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.9 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.6 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 3.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.5 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.3 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 0.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.3 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 0.7 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.8 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 0.8 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 2.1 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.3 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 1.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.2 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.7 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 1.0 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 1.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 1.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.6 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0034595 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 2.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 4.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 4.2 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.1 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 4.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.6 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 1.2 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 1.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 2.9 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.0 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.5 | GO:0019905 | syntaxin binding(GO:0019905) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.2 | 0.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 0.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 3.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 0.8 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 1.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.9 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.9 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.3 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.4 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.0 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 1.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 2.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 0.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 1.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 0.8 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.4 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 1.0 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.0 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.5 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 0.6 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 0.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 3.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 2.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.5 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.6 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 0.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |