Project
DANIO-CODE
Navigation
Downloads
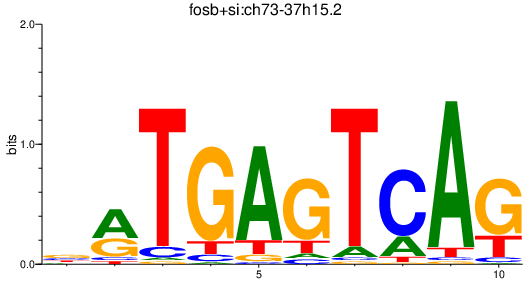
Results for fosb+si:ch73-37h15.2
Z-value: 0.37
Transcription factors associated with fosb+si:ch73-37h15.2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
fosb
|
ENSDARG00000055751 | FBJ murine osteosarcoma viral oncogene homolog B |
|
si_ch73-37h15.2
|
ENSDARG00000094107 | si_ch73-37h15.2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:ch73-37h15.2 | dr10_dc_chr5_+_62595346_62595370 | 0.40 | 1.2e-01 | Click! |
Activity profile of fosb+si:ch73-37h15.2 motif
Sorted Z-values of fosb+si:ch73-37h15.2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of fosb+si:ch73-37h15.2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_15536640 | 1.49 |
ENSDART00000098970
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr1_-_44628278 | 0.76 |
ENSDART00000133572
|
si:ch73-90k17.1
|
si:ch73-90k17.1 |
| chr6_+_44199738 | 0.74 |
ENSDART00000075486
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr24_+_19270877 | 0.74 |
|
|
|
| chr10_-_44713495 | 0.74 |
ENSDART00000076084
|
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr11_+_37371675 | 0.71 |
ENSDART00000140502
|
sh2d5
|
SH2 domain containing 5 |
| chr19_-_27966525 | 0.69 |
ENSDART00000142313
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr6_+_51713264 | 0.63 |
ENSDART00000146281
|
fam65c
|
family with sequence similarity 65, member C |
| chr7_+_38408691 | 0.59 |
ENSDART00000171691
|
psmc3
|
proteasome 26S subunit, ATPase 3 |
| chr1_-_30299017 | 0.53 |
|
|
|
| chr17_+_22291644 | 0.53 |
ENSDART00000151929
ENSDART00000089919 ENSDART00000000804 |
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr7_-_24604255 | 0.51 |
ENSDART00000173920
|
adad2
|
adenosine deaminase domain containing 2 |
| chr14_-_36004908 | 0.50 |
ENSDART00000077823
|
lrit3a
|
info leucine-rich repeat, immunoglobulin-like and transmembrane domains 3a |
| chr5_+_6476950 | 0.49 |
ENSDART00000176805
|
CABZ01112925.1
|
ENSDARG00000108220 |
| chr7_-_20358730 | 0.49 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr5_-_18393623 | 0.48 |
ENSDART00000010101
ENSDART00000099434 |
aacs
|
acetoacetyl-CoA synthetase |
| chr23_-_37010705 | 0.46 |
ENSDART00000134461
|
zgc:193690
|
zgc:193690 |
| chr1_-_29958364 | 0.46 |
ENSDART00000085454
|
dis3
|
DIS3 exosome endoribonuclease and 3'-5' exoribonuclease |
| chr23_+_25929969 | 0.45 |
ENSDART00000145426
|
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr2_+_19588034 | 0.44 |
ENSDART00000163875
|
cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr23_-_33812361 | 0.43 |
ENSDART00000136386
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr9_+_18821640 | 0.42 |
ENSDART00000006514
|
gtf2f2b
|
general transcription factor IIF, polypeptide 2b |
| chr8_+_41004169 | 0.41 |
|
|
|
| chr22_+_24194444 | 0.41 |
ENSDART00000165380
|
uchl5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr16_+_19831573 | 0.40 |
ENSDART00000135359
|
macc1
|
metastasis associated in colon cancer 1 |
| chr5_+_58936399 | 0.40 |
|
|
|
| chr15_-_23508333 | 0.40 |
ENSDART00000170427
|
ube4a
|
ubiquitination factor E4A (UFD2 homolog, yeast) |
| chr21_+_13136338 | 0.40 |
ENSDART00000142569
|
specc1lb
|
sperm antigen with calponin homology and coiled-coil domains 1-like b |
| chr5_+_33949689 | 0.40 |
ENSDART00000139317
|
aif1l
|
allograft inflammatory factor 1-like |
| chr19_+_10742944 | 0.39 |
ENSDART00000165653
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr8_-_22720007 | 0.39 |
|
|
|
| chr2_-_39026651 | 0.39 |
ENSDART00000114085
|
si:ch211-119o8.6
|
si:ch211-119o8.6 |
| chr2_-_17443607 | 0.38 |
ENSDART00000136207
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr1_-_51863300 | 0.38 |
ENSDART00000004233
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr6_-_6818607 | 0.38 |
ENSDART00000151822
|
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr6_-_31380261 | 0.38 |
|
|
|
| chr21_+_11686037 | 0.38 |
ENSDART00000031786
|
glrx
|
glutaredoxin (thioltransferase) |
| chr20_-_50089784 | 0.38 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr2_+_15432208 | 0.38 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr3_-_43253952 | 0.37 |
ENSDART00000167159
|
ubfd1
|
ubiquitin family domain containing 1 |
| chr23_-_20084191 | 0.37 |
ENSDART00000089999
|
b4galt3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr2_+_41909094 | 0.37 |
ENSDART00000179428
|
CABZ01014197.2
|
ENSDARG00000107429 |
| chr14_-_23811519 | 0.36 |
ENSDART00000124255
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr10_-_44713414 | 0.36 |
ENSDART00000076084
|
npm2b
|
nucleophosmin/nucleoplasmin, 2b |
| chr7_-_20358696 | 0.36 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr24_+_39769129 | 0.36 |
ENSDART00000066500
ENSDART00000145075 |
stub1
|
STIP1 homology and U-Box containing protein 1 |
| chr17_+_22291546 | 0.36 |
ENSDART00000151929
ENSDART00000089919 ENSDART00000000804 |
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr7_+_17848688 | 0.35 |
ENSDART00000055810
|
rab1ba
|
zRAB1B, member RAS oncogene family a |
| chr10_-_20524937 | 0.35 |
|
|
|
| chr2_+_15432130 | 0.35 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr5_+_23585615 | 0.35 |
ENSDART00000135083
|
tp53
|
tumor protein p53 |
| chr7_-_5253028 | 0.35 |
ENSDART00000033316
|
vangl2
|
VANGL planar cell polarity protein 2 |
| chr5_-_28931727 | 0.35 |
ENSDART00000174697
|
arrdc1a
|
arrestin domain containing 1a |
| chr8_-_22266750 | 0.35 |
ENSDART00000100042
ENSDART00000138966 |
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr19_-_12158958 | 0.35 |
ENSDART00000024193
|
rnf19a
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr4_+_25192397 | 0.35 |
ENSDART00000078529
ENSDART00000136643 |
kin
|
Kin17 DNA and RNA binding protein |
| chr8_-_22267056 | 0.34 |
ENSDART00000100046
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr2_+_11335810 | 0.33 |
ENSDART00000055737
|
tyw3
|
tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) |
| chr19_+_33360759 | 0.33 |
ENSDART00000052091
|
atp6v1c1b
|
ATPase, H+ transporting, lysosomal, V1 subunit C1b |
| chr7_+_56076854 | 0.33 |
|
|
|
| chr23_+_33792145 | 0.32 |
ENSDART00000024695
|
dazap2
|
DAZ associated protein 2 |
| chr16_-_24916921 | 0.32 |
ENSDART00000153731
|
si:dkey-79d12.5
|
si:dkey-79d12.5 |
| chr10_-_1933761 | 0.32 |
ENSDART00000101023
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr19_-_44161951 | 0.32 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr25_+_29752983 | 0.30 |
ENSDART00000141167
|
hsd17b12a
|
hydroxysteroid (17-beta) dehydrogenase 12a |
| chr2_-_17443642 | 0.30 |
ENSDART00000136207
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr6_-_8008902 | 0.30 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr24_-_38304083 | 0.30 |
ENSDART00000109975
|
crp7
|
C-reactive protein 7 |
| chr16_+_46443800 | 0.29 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr5_+_26888419 | 0.29 |
ENSDART00000123635
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr24_-_21224371 | 0.29 |
ENSDART00000156537
|
spice1
|
spindle and centriole associated protein 1 |
| chr20_+_23707789 | 0.29 |
|
|
|
| chr23_+_28213261 | 0.29 |
ENSDART00000143362
|
BX663515.1
|
ENSDARG00000094837 |
| chr10_+_16543480 | 0.28 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr5_+_33949469 | 0.28 |
ENSDART00000136787
|
aif1l
|
allograft inflammatory factor 1-like |
| chr5_-_71605608 | 0.28 |
ENSDART00000172302
|
wbp1
|
WW domain binding protein 1 |
| chr8_-_23316046 | 0.28 |
|
|
|
| chr5_+_44204661 | 0.28 |
ENSDART00000122288
|
ctsla
|
cathepsin La |
| chr4_-_14916491 | 0.28 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr10_-_44441390 | 0.28 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr4_+_30530 | 0.28 |
ENSDART00000157825
|
syn3
|
synapsin III |
| chr3_-_29430586 | 0.28 |
|
|
|
| chr11_-_11925801 | 0.27 |
|
|
|
| chr24_-_21224397 | 0.27 |
ENSDART00000156537
|
spice1
|
spindle and centriole associated protein 1 |
| chr3_-_36126124 | 0.27 |
|
|
|
| chr3_-_20944854 | 0.27 |
ENSDART00000109790
|
nlk1
|
nemo-like kinase, type 1 |
| chr8_-_22267016 | 0.27 |
ENSDART00000140978
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr2_+_11335733 | 0.26 |
ENSDART00000055737
|
tyw3
|
tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) |
| chr22_-_26254136 | 0.26 |
ENSDART00000060978
|
wdr83
|
WD repeat domain containing 83 |
| chr8_-_23316108 | 0.26 |
|
|
|
| chr8_-_1586037 | 0.26 |
ENSDART00000142703
|
CU694486.1
|
ENSDARG00000091948 |
| chr9_+_33344121 | 0.26 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr7_-_5252575 | 0.26 |
|
|
|
| chr10_-_25737220 | 0.25 |
ENSDART00000135058
|
sod1
|
superoxide dismutase 1, soluble |
| chr18_+_44856321 | 0.25 |
ENSDART00000097328
|
arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr1_-_46430811 | 0.25 |
ENSDART00000048445
|
mhc1zea
|
major histocompatibility complex class I ZEA |
| chr2_-_30012325 | 0.25 |
ENSDART00000020792
|
cnpy1
|
canopy1 |
| chr23_+_25930072 | 0.25 |
ENSDART00000124103
|
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr15_+_36371548 | 0.24 |
|
|
|
| chr1_-_44628181 | 0.24 |
ENSDART00000133572
|
si:ch73-90k17.1
|
si:ch73-90k17.1 |
| chr24_-_20946041 | 0.24 |
ENSDART00000140786
|
qtrtd1
|
queuine tRNA-ribosyltransferase domain containing 1 |
| chr7_+_26378093 | 0.24 |
ENSDART00000173823
ENSDART00000101053 |
tp53i11a
|
tumor protein p53 inducible protein 11a |
| chr23_-_20084226 | 0.23 |
ENSDART00000163396
|
b4galt3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr21_-_13759752 | 0.23 |
ENSDART00000102197
|
fam129ba
|
family with sequence similarity 129, member Ba |
| chr11_+_11284030 | 0.23 |
ENSDART00000026814
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr3_+_23963926 | 0.22 |
ENSDART00000138270
|
copz2
|
coatomer protein complex, subunit zeta 2 |
| chr3_+_48319803 | 0.22 |
ENSDART00000113531
|
mkl2b
|
MKL/myocardin-like 2b |
| chr2_+_11335970 | 0.22 |
ENSDART00000055737
|
tyw3
|
tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) |
| chr7_-_20358897 | 0.22 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr22_-_9832436 | 0.22 |
|
|
|
| chr5_+_56896961 | 0.22 |
|
|
|
| chr8_-_22266835 | 0.22 |
ENSDART00000138966
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr12_+_46281592 | 0.21 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr25_+_3423586 | 0.21 |
ENSDART00000166312
|
ccdc77
|
coiled-coil domain containing 77 |
| chr14_-_46599151 | 0.21 |
|
|
|
| chr25_-_22094023 | 0.21 |
ENSDART00000067478
|
pkp3a
|
plakophilin 3a |
| chr14_+_21522552 | 0.21 |
ENSDART00000122739
|
ctbp1
|
C-terminal binding protein 1 |
| chr2_+_11336033 | 0.21 |
ENSDART00000055737
|
tyw3
|
tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) |
| chr10_+_42841562 | 0.21 |
|
|
|
| chr23_+_5631846 | 0.20 |
ENSDART00000059307
|
smpd2a
|
sphingomyelin phosphodiesterase 2a, neutral membrane (neutral sphingomyelinase) |
| chr10_+_2147867 | 0.20 |
ENSDART00000143636
|
cntnap3
|
contactin associated protein-like 3 |
| chr13_-_26668719 | 0.20 |
ENSDART00000139264
|
vrk2
|
vaccinia related kinase 2 |
| chr5_+_44204845 | 0.20 |
ENSDART00000141198
|
ctsla
|
cathepsin La |
| chr15_-_26153718 | 0.20 |
ENSDART00000140214
|
dph1
|
diphthamide biosynthesis 1 |
| chr17_-_43725091 | 0.20 |
|
|
|
| chr19_+_7082199 | 0.20 |
ENSDART00000110366
|
zbtb22b
|
zinc finger and BTB domain containing 22b |
| chr7_-_20358841 | 0.19 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr7_-_31346844 | 0.19 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr24_-_33475705 | 0.19 |
ENSDART00000074161
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr1_-_39622531 | 0.19 |
|
|
|
| chr17_-_23201467 | 0.19 |
ENSDART00000156411
|
fam98a
|
family with sequence similarity 98, member A |
| chr25_-_14282280 | 0.19 |
ENSDART00000043812
ENSDART00000144414 |
polr2c
|
polymerase (RNA) II (DNA directed) polypeptide C |
| chr13_-_24695520 | 0.18 |
ENSDART00000142745
|
slka
|
STE20-like kinase a |
| chr21_+_11685992 | 0.18 |
ENSDART00000031786
|
glrx
|
glutaredoxin (thioltransferase) |
| chr5_+_44205128 | 0.18 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr4_-_4526152 | 0.18 |
ENSDART00000057519
|
zgc:194209
|
zgc:194209 |
| chr10_-_44441197 | 0.18 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr15_+_23508351 | 0.18 |
ENSDART00000162997
|
phykpl
|
5-phosphohydroxy-L-lysine phospho-lyase |
| chr24_+_39768911 | 0.18 |
ENSDART00000066500
ENSDART00000145075 |
stub1
|
STIP1 homology and U-Box containing protein 1 |
| chr10_+_187740 | 0.18 |
ENSDART00000167367
|
ets2
|
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
| chr7_+_26730804 | 0.17 |
|
|
|
| chr13_+_20742320 | 0.17 |
|
|
|
| chr6_-_47928548 | 0.17 |
|
|
|
| chr23_-_37010664 | 0.17 |
ENSDART00000102886
|
zgc:193690
|
zgc:193690 |
| chr15_+_17407036 | 0.17 |
ENSDART00000018461
|
vmp1
|
vacuole membrane protein 1 |
| chr3_+_25868700 | 0.17 |
ENSDART00000024316
|
mcm5
|
minichromosome maintenance complex component 5 |
| chr8_-_7584890 | 0.17 |
ENSDART00000137975
|
irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr4_-_7644310 | 0.17 |
ENSDART00000157751
|
BX649411.1
|
ENSDARG00000101254 |
| chr8_+_21974863 | 0.17 |
|
|
|
| chr12_+_46281623 | 0.17 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr10_-_20524860 | 0.16 |
|
|
|
| chr12_+_23250339 | 0.16 |
ENSDART00000078896
|
waca
|
WW domain containing adaptor with coiled-coil a |
| chr2_+_19588085 | 0.16 |
ENSDART00000163875
|
cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr17_+_990022 | 0.16 |
ENSDART00000169903
|
CABZ01060373.1
|
ENSDARG00000103671 |
| chr25_-_16685648 | 0.16 |
ENSDART00000019413
|
ndufa9a
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9a |
| chr14_-_5509554 | 0.16 |
|
|
|
| chr12_+_46281511 | 0.15 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr3_-_34687314 | 0.15 |
ENSDART00000084448
|
psmd11a
|
proteasome 26S subunit, non-ATPase 11a |
| chr5_-_31889551 | 0.15 |
ENSDART00000168870
|
gpr107
|
G protein-coupled receptor 107 |
| chr6_+_20008821 | 0.15 |
ENSDART00000151205
|
aldh1l1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr13_+_46487890 | 0.15 |
|
|
|
| chr6_-_37766424 | 0.15 |
ENSDART00000149722
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr11_+_5638359 | 0.14 |
|
|
|
| chr5_+_58936508 | 0.14 |
|
|
|
| chr21_+_11685911 | 0.14 |
ENSDART00000031786
|
glrx
|
glutaredoxin (thioltransferase) |
| chr23_-_9872524 | 0.14 |
ENSDART00000133602
|
mapre1b
|
microtubule-associated protein, RP/EB family, member 1b |
| chr19_+_42863138 | 0.14 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr21_-_20983956 | 0.14 |
ENSDART00000064032
|
eif4ebp1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr16_+_24045675 | 0.14 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr8_-_20213787 | 0.13 |
ENSDART00000136911
|
acer1
|
alkaline ceramidase 1 |
| chr21_+_22856217 | 0.13 |
ENSDART00000065565
|
alg8
|
ALG8, alpha-1,3-glucosyltransferase |
| chr25_+_29752909 | 0.13 |
ENSDART00000005299
|
hsd17b12a
|
hydroxysteroid (17-beta) dehydrogenase 12a |
| chr8_-_53593850 | 0.13 |
|
|
|
| chr6_+_12091402 | 0.13 |
ENSDART00000155101
|
si:dkey-276j7.3
|
si:dkey-276j7.3 |
| chr15_-_31296524 | 0.13 |
ENSDART00000008854
|
wsb1
|
WD repeat and SOCS box containing 1 |
| chr8_-_10912137 | 0.13 |
ENSDART00000064058
ENSDART00000162109 ENSDART00000101277 |
pqlc2
|
PQ loop repeat containing 2 |
| chr9_+_426701 | 0.13 |
ENSDART00000164310
|
bzw1b
|
basic leucine zipper and W2 domains 1b |
| chr3_-_20944652 | 0.13 |
ENSDART00000109790
|
nlk1
|
nemo-like kinase, type 1 |
| chr6_-_7625286 | 0.13 |
|
|
|
| chr12_+_25341625 | 0.13 |
ENSDART00000011662
|
ppm1bb
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Bb |
| chr16_+_24045707 | 0.13 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr23_+_24575330 | 0.12 |
ENSDART00000078824
|
szrd1
|
SUZ RNA binding domain containing 1 |
| chr15_-_31296457 | 0.12 |
ENSDART00000008854
|
wsb1
|
WD repeat and SOCS box containing 1 |
| chr8_+_17975195 | 0.12 |
ENSDART00000039887
ENSDART00000121984 |
ssbp3b
|
single stranded DNA binding protein 3b |
| chr15_-_1928150 | 0.12 |
|
|
|
| chr12_+_25341263 | 0.12 |
ENSDART00000153333
|
ppm1bb
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Bb |
| chr16_+_30168944 | 0.12 |
|
|
|
| chr16_-_54679352 | 0.11 |
ENSDART00000083713
|
clk2b
|
CDC-like kinase 2b |
| chr14_+_21522672 | 0.11 |
ENSDART00000122739
|
ctbp1
|
C-terminal binding protein 1 |
| chr1_+_40784540 | 0.11 |
ENSDART00000142508
|
si:ch211-89o9.6
|
si:ch211-89o9.6 |
| chr4_+_76548332 | 0.11 |
ENSDART00000159452
ENSDART00000092415 |
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr12_-_3042573 | 0.11 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr23_+_33792005 | 0.11 |
ENSDART00000024695
|
dazap2
|
DAZ associated protein 2 |
| chr8_-_53593880 | 0.11 |
ENSDART00000015554
|
ubiad1
|
UbiA prenyltransferase domain containing 1 |
| chr12_-_19056530 | 0.11 |
|
|
|
| chr20_+_23707625 | 0.11 |
|
|
|
| chr22_+_39012456 | 0.10 |
ENSDART00000176985
|
CABZ01044615.1
|
ENSDARG00000108810 |
| chr22_+_3989571 | 0.10 |
ENSDART00000170751
|
ticam1
|
toll-like receptor adaptor molecule 1 |
| chr17_-_20675997 | 0.10 |
ENSDART00000063613
ENSDART00000171575 |
ccdc6a
|
coiled-coil domain containing 6a |
| chr9_+_33343936 | 0.10 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 0.5 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.2 | 0.5 | GO:0046887 | positive regulation of peptide secretion(GO:0002793) positive regulation of insulin secretion(GO:0032024) positive regulation of hormone secretion(GO:0046887) positive regulation of peptide hormone secretion(GO:0090277) |
| 0.1 | 0.3 | GO:0045813 | positive regulation of Wnt signaling pathway, calcium modulating pathway(GO:0045813) |
| 0.1 | 0.3 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.1 | 0.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.4 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.1 | 0.9 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.3 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.5 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) regulation of steroid metabolic process(GO:0019218) |
| 0.1 | 0.4 | GO:0008210 | estrogen biosynthetic process(GO:0006703) estrogen metabolic process(GO:0008210) |
| 0.1 | 0.7 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.1 | 0.6 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.5 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.5 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.7 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 1.1 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.1 | 0.2 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.7 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.4 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.6 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.2 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.2 | GO:0032197 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.3 | GO:0019430 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.3 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.2 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 1.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.1 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.3 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.4 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.0 | 0.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.1 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.4 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.2 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.2 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.1 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.4 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.1 | GO:0021592 | fourth ventricle development(GO:0021592) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 0.4 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.5 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.4 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.4 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.7 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.9 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.3 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 0.4 | GO:0090624 | endoribonuclease activity, cleaving miRNA-paired mRNA(GO:0090624) |
| 0.1 | 0.2 | GO:0016844 | amine-lyase activity(GO:0016843) strictosidine synthase activity(GO:0016844) |
| 0.1 | 0.4 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 0.3 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.3 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 1.0 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.4 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.5 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 0.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.0 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.4 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.1 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |