Project
DANIO-CODE
Navigation
Downloads
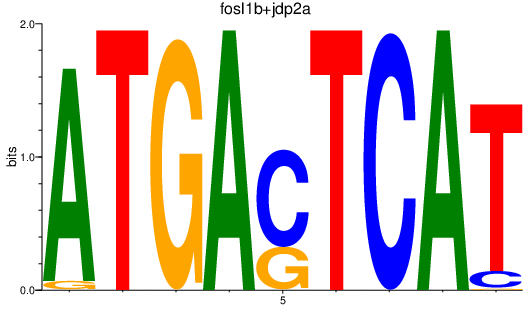
Results for fosl1b+jdp2a
Z-value: 1.13
Transcription factors associated with fosl1b+jdp2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
jdp2a
|
ENSDARG00000040137 | Jun dimerization protein 2a |
|
fosl1b
|
ENSDARG00000079373 | FOS-like antigen 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| jdp2a | dr10_dc_chr17_-_50140768_50140778 | -0.42 | 1.0e-01 | Click! |
Activity profile of fosl1b+jdp2a motif
Sorted Z-values of fosl1b+jdp2a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of fosl1b+jdp2a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_30560440 | 3.88 |
ENSDART00000079068
|
asb11
|
ankyrin repeat and SOCS box containing 11 |
| chr3_+_24067387 | 2.96 |
ENSDART00000055609
|
atf4b
|
activating transcription factor 4b |
| chr8_-_38168395 | 2.60 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr21_-_2147438 | 2.52 |
ENSDART00000172276
|
zgc:163077
|
zgc:163077 |
| chr8_+_25880895 | 2.31 |
ENSDART00000124300
|
rhoab
|
ras homolog gene family, member Ab |
| chr21_+_25729090 | 2.27 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr11_-_5793 | 2.25 |
|
|
|
| chr19_+_17451381 | 2.15 |
ENSDART00000167602
|
spaca4l
|
sperm acrosome associated 4 like |
| chr22_+_16471319 | 2.14 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr23_+_25930072 | 2.13 |
ENSDART00000124103
|
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr25_-_18234069 | 2.10 |
ENSDART00000104496
|
dusp6
|
dual specificity phosphatase 6 |
| chr20_+_30948175 | 1.93 |
|
|
|
| chr4_+_56996802 | 1.81 |
|
|
|
| chr11_-_11925832 | 1.78 |
|
|
|
| chr23_+_116397 | 1.75 |
|
|
|
| chr25_-_22089794 | 1.49 |
ENSDART00000139110
|
pkp3a
|
plakophilin 3a |
| chr14_-_40454194 | 1.42 |
ENSDART00000166621
|
ef1
|
E74-like factor 1 (ets domain transcription factor) |
| chr11_-_18649022 | 1.42 |
|
|
|
| chr8_-_337744 | 1.39 |
|
|
|
| chr16_-_31517906 | 1.36 |
ENSDART00000145691
|
ENSDARG00000054814
|
ENSDARG00000054814 |
| chr6_-_47928548 | 1.29 |
|
|
|
| chr19_-_7576069 | 1.25 |
ENSDART00000148836
|
rfx5
|
regulatory factor X, 5 |
| chr13_-_3801122 | 1.24 |
|
|
|
| chr19_-_19888074 | 1.20 |
|
|
|
| chr4_-_71353374 | 1.20 |
|
|
|
| chr5_-_30324182 | 1.20 |
ENSDART00000153909
|
spns2
|
spinster homolog 2 (Drosophila) |
| chr8_+_46319434 | 1.15 |
ENSDART00000145618
|
si:dkey-75a21.2
|
si:dkey-75a21.2 |
| chr21_-_2147085 | 1.11 |
ENSDART00000169262
|
zgc:163077
|
zgc:163077 |
| chr4_-_49071258 | 1.08 |
ENSDART00000162560
|
BX855613.1
|
ENSDARG00000098743 |
| chr6_-_47928632 | 1.04 |
|
|
|
| chr20_+_28465622 | 1.03 |
ENSDART00000146905
ENSDART00000103355 |
rhov
|
ras homolog family member V |
| chr20_-_26632676 | 1.01 |
ENSDART00000131994
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr5_+_23585492 | 1.00 |
ENSDART00000135083
|
tp53
|
tumor protein p53 |
| chr4_-_6800721 | 0.99 |
ENSDART00000099467
|
ifrd1
|
interferon-related developmental regulator 1 |
| chr16_+_42925950 | 0.94 |
ENSDART00000159730
ENSDART00000014956 |
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr17_+_28657998 | 0.94 |
ENSDART00000159067
|
hectd1
|
HECT domain containing 1 |
| chr21_-_36338514 | 0.94 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr13_+_7243353 | 0.92 |
ENSDART00000146592
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr6_+_42478185 | 0.86 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr21_+_22808694 | 0.85 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr15_-_40391882 | 0.81 |
|
|
|
| chr5_+_41680352 | 0.80 |
ENSDART00000142855
|
tbx6l
|
T-box 6, like |
| chr23_-_30119058 | 0.79 |
ENSDART00000103480
|
ccdc187
|
coiled-coil domain containing 187 |
| chr14_-_5509554 | 0.75 |
|
|
|
| KN149695v1_+_67009 | 0.74 |
ENSDART00000178024
|
ENSDARG00000107779
|
ENSDARG00000107779 |
| chr4_-_75690772 | 0.73 |
ENSDART00000174183
|
CABZ01063884.1
|
ENSDARG00000105697 |
| chr11_+_26152136 | 0.70 |
|
|
|
| chr7_+_5408319 | 0.70 |
ENSDART00000142709
|
si:dkeyp-67a8.4
|
si:dkeyp-67a8.4 |
| chr21_-_2222412 | 0.68 |
ENSDART00000169448
|
si:ch73-299h12.1
|
si:ch73-299h12.1 |
| chr4_-_36838449 | 0.68 |
|
|
|
| chr3_-_44113070 | 0.67 |
ENSDART00000160717
|
znf750
|
zinc finger protein 750 |
| chr4_+_52851409 | 0.63 |
|
|
|
| chr19_-_10962536 | 0.61 |
ENSDART00000160438
|
psmd4a
|
proteasome 26S subunit, non-ATPase 4a |
| chr3_+_30790513 | 0.60 |
ENSDART00000130422
|
cldni
|
claudin i |
| chr16_-_31517858 | 0.59 |
ENSDART00000145691
|
ENSDARG00000054814
|
ENSDARG00000054814 |
| chr8_+_48976657 | 0.57 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr5_+_19506512 | 0.56 |
ENSDART00000047841
|
sgsm1a
|
small G protein signaling modulator 1a |
| chr7_+_38455040 | 0.55 |
|
|
|
| chr5_+_40910195 | 0.53 |
ENSDART00000134317
|
vcp
|
valosin containing protein |
| chr8_+_48976994 | 0.52 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr20_+_25726978 | 0.51 |
ENSDART00000164264
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr5_+_15319430 | 0.51 |
ENSDART00000162003
|
hspb8
|
heat shock protein b8 |
| chr21_-_36338704 | 0.50 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr23_+_25929969 | 0.49 |
ENSDART00000145426
|
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr22_+_39098205 | 0.49 |
ENSDART00000002826
|
gmppb
|
GDP-mannose pyrophosphorylase B |
| chr7_+_23306546 | 0.47 |
ENSDART00000049885
|
si:dkey-172j4.3
|
si:dkey-172j4.3 |
| chr13_+_35213326 | 0.46 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr13_+_1051015 | 0.46 |
ENSDART00000033528
|
tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr13_+_46487890 | 0.45 |
|
|
|
| chr4_+_56143439 | 0.45 |
ENSDART00000178597
|
si:ch211-212k5.1
|
si:ch211-212k5.1 |
| chr20_-_16649213 | 0.44 |
ENSDART00000137601
|
ches1
|
checkpoint suppressor 1 |
| chr22_-_36718623 | 0.43 |
|
|
|
| chr12_+_37005712 | 0.43 |
|
|
|
| chr17_+_23536720 | 0.43 |
ENSDART00000143508
|
pank1a
|
pantothenate kinase 1a |
| chr9_-_12913920 | 0.42 |
ENSDART00000122799
|
ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr13_-_36996246 | 0.42 |
ENSDART00000133242
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr6_-_49064365 | 0.41 |
ENSDART00000156124
|
si:ch211-105j21.9
|
si:ch211-105j21.9 |
| KN149946v1_-_12861 | 0.41 |
ENSDART00000163016
|
CABZ01048402.2
|
ENSDARG00000103875 |
| chr3_+_19066717 | 0.40 |
ENSDART00000134433
|
il12rb2l
|
interleukin 12 receptor, beta 2a, like |
| chr25_+_29752909 | 0.40 |
ENSDART00000005299
|
hsd17b12a
|
hydroxysteroid (17-beta) dehydrogenase 12a |
| chr4_+_46414646 | 0.40 |
ENSDART00000179615
|
BX324155.2
|
ENSDARG00000107359 |
| chr5_-_4082749 | 0.39 |
|
|
|
| chr13_+_7243325 | 0.39 |
ENSDART00000146592
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr11_-_269181 | 0.39 |
|
|
|
| KN149726v1_+_1094 | 0.37 |
|
|
|
| chr14_+_50394381 | 0.36 |
ENSDART00000158329
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr11_+_37639045 | 0.36 |
ENSDART00000111157
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr20_+_25726732 | 0.36 |
ENSDART00000078385
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr16_-_27609082 | 0.36 |
|
|
|
| chr18_+_45680493 | 0.35 |
ENSDART00000109948
|
qser1
|
glutamine and serine rich 1 |
| chr5_+_17120453 | 0.34 |
|
|
|
| chr6_+_56157608 | 0.34 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr4_+_4825409 | 0.33 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr8_-_31597252 | 0.32 |
ENSDART00000018886
|
ghra
|
growth hormone receptor a |
| chr6_-_31380261 | 0.31 |
|
|
|
| chr1_+_15827083 | 0.31 |
|
|
|
| chr12_-_20493683 | 0.30 |
|
|
|
| chr1_+_52365033 | 0.30 |
ENSDART00000133411
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr18_+_21419735 | 0.28 |
ENSDART00000144523
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| chr25_-_10930008 | 0.28 |
ENSDART00000156846
|
mespbb
|
mesoderm posterior bb |
| chr14_+_6308806 | 0.28 |
ENSDART00000139292
|
si:dkeyp-44a8.2
|
si:dkeyp-44a8.2 |
| chr19_-_19888009 | 0.28 |
|
|
|
| chr16_+_13964949 | 0.28 |
ENSDART00000143983
|
zgc:174888
|
zgc:174888 |
| chr3_-_16569378 | 0.27 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr8_+_48976926 | 0.27 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr14_-_25636795 | 0.27 |
|
|
|
| KN150708v1_+_30684 | 0.26 |
|
|
|
| chr7_-_29262814 | 0.26 |
ENSDART00000124028
|
anxa2b
|
annexin A2b |
| chr20_+_32575184 | 0.26 |
ENSDART00000145269
|
ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr25_+_16215855 | 0.25 |
ENSDART00000064187
|
parvaa
|
parvin, alpha a |
| chr7_+_6786457 | 0.25 |
ENSDART00000172421
|
rbm14b
|
RNA binding motif protein 14b |
| chr5_-_62963536 | 0.24 |
ENSDART00000050865
|
surf4l
|
surfeit gene 4, like |
| chr20_+_49975268 | 0.24 |
ENSDART00000008808
|
hmbox1b
|
homeobox containing 1 b |
| chr2_-_23310316 | 0.23 |
ENSDART00000146014
|
sap130b
|
Sin3A-associated protein b |
| chr7_+_71262935 | 0.23 |
ENSDART00000100396
|
ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr7_-_25426346 | 0.23 |
ENSDART00000082620
|
dysf
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) |
| chr22_+_11505448 | 0.23 |
ENSDART00000113930
|
npb
|
neuropeptide B |
| chr5_+_37184963 | 0.23 |
ENSDART00000053511
|
myo1ca
|
myosin Ic, paralog a |
| chr22_+_9832516 | 0.23 |
ENSDART00000105942
|
si:dkey-253d23.3
|
si:dkey-253d23.3 |
| chr5_-_19691538 | 0.22 |
ENSDART00000139675
|
dao.1
|
D-amino-acid oxidase, tandem duplicate 1 |
| chr24_+_22195227 | 0.22 |
|
|
|
| chr16_-_13927684 | 0.22 |
ENSDART00000146719
|
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr11_+_268997 | 0.22 |
ENSDART00000168188
|
mettl1
|
methyltransferase like 1 |
| chr3_-_15584548 | 0.22 |
ENSDART00000137325
|
mvp
|
major vault protein |
| chr4_-_12782246 | 0.22 |
ENSDART00000058020
|
helb
|
helicase (DNA) B |
| chr20_-_40823307 | 0.21 |
ENSDART00000061261
|
cx43
|
connexin 43 |
| chr18_+_13108810 | 0.21 |
ENSDART00000161153
|
GAN
|
gigaxonin |
| chr11_-_25223723 | 0.21 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr13_+_46899292 | 0.20 |
|
|
|
| chr9_-_30560373 | 0.20 |
ENSDART00000079068
|
asb11
|
ankyrin repeat and SOCS box containing 11 |
| chr9_-_2416300 | 0.20 |
|
|
|
| chr13_-_24779651 | 0.20 |
ENSDART00000077775
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr25_+_29752983 | 0.19 |
ENSDART00000141167
|
hsd17b12a
|
hydroxysteroid (17-beta) dehydrogenase 12a |
| chr1_-_39622531 | 0.19 |
|
|
|
| chr16_-_43090120 | 0.19 |
ENSDART00000155637
|
BX005234.1
|
ENSDARG00000097761 |
| chr16_-_13927947 | 0.19 |
ENSDART00000144856
|
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr25_+_18487313 | 0.18 |
ENSDART00000148741
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr13_-_37523664 | 0.18 |
ENSDART00000115354
|
si:dkey-188i13.10
|
si:dkey-188i13.10 |
| chr23_-_1008307 | 0.17 |
ENSDART00000110588
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr3_+_25933164 | 0.17 |
ENSDART00000143697
|
si:dkeyp-69e1.8
|
si:dkeyp-69e1.8 |
| chr21_-_18787657 | 0.17 |
|
|
|
| chr1_+_19376127 | 0.16 |
ENSDART00000088653
|
prss12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr17_-_5426195 | 0.16 |
ENSDART00000035944
|
clic5a
|
chloride intracellular channel 5a |
| chr11_+_6118826 | 0.16 |
ENSDART00000159680
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| KN149735v1_-_1679 | 0.16 |
|
|
|
| chr21_+_26576104 | 0.16 |
ENSDART00000142667
|
esrra
|
estrogen-related receptor alpha |
| chr5_+_33949469 | 0.15 |
ENSDART00000136787
|
aif1l
|
allograft inflammatory factor 1-like |
| chr15_+_17407258 | 0.15 |
ENSDART00000111753
|
vmp1
|
vacuole membrane protein 1 |
| chr3_-_39245184 | 0.15 |
|
|
|
| chr23_-_9872524 | 0.15 |
ENSDART00000133602
|
mapre1b
|
microtubule-associated protein, RP/EB family, member 1b |
| chr12_-_36113358 | 0.15 |
|
|
|
| chr10_+_35479226 | 0.15 |
ENSDART00000147303
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr22_-_20551624 | 0.15 |
|
|
|
| chr4_+_4825628 | 0.15 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr16_-_24916921 | 0.14 |
ENSDART00000153731
|
si:dkey-79d12.5
|
si:dkey-79d12.5 |
| chr6_-_39313219 | 0.14 |
|
|
|
| chr10_+_5693933 | 0.14 |
ENSDART00000159769
|
pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr24_-_32750010 | 0.13 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr9_+_22846535 | 0.13 |
ENSDART00000101765
|
si:dkey-189g17.2
|
si:dkey-189g17.2 |
| chr3_+_24067430 | 0.12 |
ENSDART00000055609
|
atf4b
|
activating transcription factor 4b |
| chr22_-_36718332 | 0.12 |
|
|
|
| chr21_-_11564398 | 0.12 |
ENSDART00000141297
|
cast
|
calpastatin |
| chr19_-_29706969 | 0.11 |
ENSDART00000130815
ENSDART00000172590 ENSDART00000103437 |
e2f3
|
E2F transcription factor 3 |
| chr14_+_34146377 | 0.11 |
ENSDART00000131861
|
tmsb2
|
thymosin beta 2 |
| chr8_-_11132386 | 0.11 |
ENSDART00000133532
|
ENSDARG00000078221
|
ENSDARG00000078221 |
| chr6_+_49489473 | 0.11 |
ENSDART00000177749
|
FO704848.1
|
ENSDARG00000107438 |
| chr21_-_7702431 | 0.10 |
|
|
|
| chr8_-_39944817 | 0.10 |
ENSDART00000083066
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr24_-_3273409 | 0.10 |
|
|
|
| chr5_-_22981395 | 0.10 |
ENSDART00000170293
|
si:dkeyp-20g2.1
|
si:dkeyp-20g2.1 |
| chr17_+_45430353 | 0.10 |
ENSDART00000162937
|
ezra
|
ezrin a |
| chr8_+_36522231 | 0.09 |
ENSDART00000126687
|
sf3a1
|
splicing factor 3a, subunit 1 |
| chr4_-_22590638 | 0.09 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr25_+_9904821 | 0.09 |
|
|
|
| chr17_-_49931552 | 0.09 |
ENSDART00000161008
|
filip1a
|
filamin A interacting protein 1a |
| chr13_+_21757676 | 0.08 |
ENSDART00000162709
|
ndst2a
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2a |
| chr4_+_5308883 | 0.08 |
ENSDART00000150366
|
si:ch211-214j24.10
|
si:ch211-214j24.10 |
| chr14_-_33704021 | 0.08 |
ENSDART00000149396
ENSDART00000123607 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr8_-_36522230 | 0.08 |
ENSDART00000132804
|
ccdc157
|
coiled-coil domain containing 157 |
| chr5_-_19619115 | 0.06 |
ENSDART00000026516
|
pxmp2
|
peroxisomal membrane protein 2 |
| chr11_-_6004509 | 0.06 |
ENSDART00000108628
|
ano8b
|
anoctamin 8b |
| chr10_+_35478939 | 0.06 |
ENSDART00000147303
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr23_+_39569262 | 0.06 |
ENSDART00000031616
|
mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr1_-_22170553 | 0.06 |
ENSDART00000139412
|
SMIM18
|
small integral membrane protein 18 |
| chr2_-_44893769 | 0.06 |
ENSDART00000041806
|
acsm3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr5_+_50306662 | 0.05 |
ENSDART00000149892
|
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr7_+_23604092 | 0.05 |
ENSDART00000101406
|
rab39bb
|
RAB39B, member RAS oncogene family b |
| chr21_-_44570264 | 0.05 |
ENSDART00000159323
|
fundc2
|
fun14 domain containing 2 |
| chr13_+_46899421 | 0.04 |
|
|
|
| chr24_-_36412892 | 0.04 |
ENSDART00000148868
|
coasy
|
CoA synthase |
| chr12_-_29118423 | 0.04 |
ENSDART00000153175
|
si:ch211-214e3.5
|
si:ch211-214e3.5 |
| chr11_-_25223169 | 0.04 |
|
|
|
| chr16_-_34330688 | 0.03 |
ENSDART00000044235
ENSDART00000159744 |
phactr4b
|
phosphatase and actin regulator 4b |
| chr21_+_10769107 | 0.03 |
|
|
|
| chr1_-_18084246 | 0.03 |
|
|
|
| chr12_+_23250339 | 0.03 |
ENSDART00000078896
|
waca
|
WW domain containing adaptor with coiled-coil a |
| chr3_+_16826320 | 0.03 |
ENSDART00000112450
|
cavin1a
|
caveolae associated protein 1a |
| chr4_+_4825461 | 0.03 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr23_-_1008468 | 0.02 |
ENSDART00000110588
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr5_+_23585615 | 0.02 |
ENSDART00000135083
|
tp53
|
tumor protein p53 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.7 | 2.1 | GO:1905178 | regulation of cardiac muscle tissue regeneration(GO:1905178) |
| 0.5 | 2.3 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.3 | 1.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.3 | 1.0 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.2 | 0.8 | GO:0010939 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) |
| 0.2 | 2.6 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.2 | 0.5 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.2 | 1.0 | GO:1901216 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.2 | 0.5 | GO:0071947 | protein K29-linked deubiquitination(GO:0035523) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 2.2 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 1.4 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 2.9 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 1.0 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.6 | GO:0006703 | estrogen biosynthetic process(GO:0006703) estrogen metabolic process(GO:0008210) |
| 0.1 | 0.8 | GO:0009113 | purine nucleobase biosynthetic process(GO:0009113) |
| 0.1 | 0.5 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.2 | GO:0050765 | monocyte activation involved in immune response(GO:0002280) negative regulation of phagocytosis(GO:0050765) |
| 0.1 | 0.3 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 1.1 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 0.2 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 1.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.3 | GO:0060416 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.2 | GO:0070178 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.1 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.3 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.3 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.0 | 0.3 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.9 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 1.5 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.0 | 0.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.0 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.2 | GO:0033505 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.3 | GO:0071698 | olfactory placode development(GO:0071698) |
| 0.0 | 0.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.5 | GO:0034620 | cellular response to unfolded protein(GO:0034620) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.1 | 0.3 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 1.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 1.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 2.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 2.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.6 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 2.9 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.3 | 0.9 | GO:0004044 | amidophosphoribosyltransferase activity(GO:0004044) |
| 0.2 | 2.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 1.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 0.5 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 2.6 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.3 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.1 | 2.2 | GO:0004675 | transmembrane receptor protein serine/threonine kinase activity(GO:0004675) |
| 0.1 | 0.6 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 0.2 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.1 | 0.8 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 2.0 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 1.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 2.9 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.1 | GO:0016842 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) amidine-lyase activity(GO:0016842) |
| 0.0 | 0.3 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 1.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 3.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.2 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 1.2 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.4 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 1.1 | GO:0019199 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) transmembrane receptor protein kinase activity(GO:0019199) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.7 | GO:0019901 | protein kinase binding(GO:0019901) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 2.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 1.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 1.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.2 | 2.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.0 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 3.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |