Project
DANIO-CODE
Navigation
Downloads
Results for fosl2l
Z-value: 0.52
Transcription factors associated with fosl2l
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
fosl2l
|
ENSDARG00000092174 | fos-like antigen 2 like |
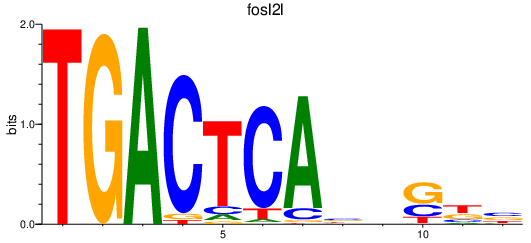
Activity profile of fosl2l motif
Sorted Z-values of fosl2l motif
Network of associatons between targets according to the STRING database.
First level regulatory network of fosl2l
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_5845303 | 1.11 |
ENSDART00000163948
|
ENSDARG00000053246
|
ENSDARG00000053246 |
| chr1_+_45907269 | 0.95 |
ENSDART00000132861
|
cab39l
|
calcium binding protein 39-like |
| chr7_+_66660882 | 0.66 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr6_+_48978653 | 0.61 |
|
|
|
| chr8_-_22267056 | 0.60 |
ENSDART00000100046
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr12_-_3042394 | 0.60 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr8_-_22266750 | 0.58 |
ENSDART00000100042
ENSDART00000138966 |
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr19_+_40558066 | 0.58 |
ENSDART00000049968
|
si:ch211-173p18.3
|
si:ch211-173p18.3 |
| chr24_-_23793117 | 0.56 |
|
|
|
| chr15_-_43954665 | 0.56 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr8_-_22267016 | 0.56 |
ENSDART00000140978
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr9_+_28329384 | 0.50 |
ENSDART00000046880
|
plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| chr1_+_45907424 | 0.49 |
ENSDART00000053240
|
cab39l
|
calcium binding protein 39-like |
| chr3_-_20912934 | 0.49 |
ENSDART00000156275
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr23_+_19728953 | 0.49 |
ENSDART00000104441
|
abhd6b
|
abhydrolase domain containing 6b |
| chr9_-_33799681 | 0.47 |
ENSDART00000158678
|
caska
|
calcium/calmodulin-dependent serine protein kinase a |
| chr24_-_21224371 | 0.43 |
ENSDART00000156537
|
spice1
|
spindle and centriole associated protein 1 |
| chr5_+_35186037 | 0.42 |
ENSDART00000127383
ENSDART00000022043 |
stard8
|
StAR-related lipid transfer (START) domain containing 8 |
| chr5_-_46028578 | 0.42 |
|
|
|
| chr19_+_37533306 | 0.42 |
ENSDART00000032341
|
pef1
|
penta-EF-hand domain containing 1 |
| chr5_-_1102450 | 0.41 |
ENSDART00000115044
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr15_+_17407036 | 0.40 |
ENSDART00000018461
|
vmp1
|
vacuole membrane protein 1 |
| chr16_+_26673760 | 0.40 |
ENSDART00000141393
|
ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr19_+_15536640 | 0.40 |
ENSDART00000098970
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr12_+_46281623 | 0.39 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr22_+_24576319 | 0.38 |
ENSDART00000164256
|
ENSDARG00000100186
|
ENSDARG00000100186 |
| chr8_-_22266835 | 0.37 |
ENSDART00000138966
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr23_+_25062876 | 0.37 |
ENSDART00000145307
ENSDART00000172299 |
arhgap4a
|
Rho GTPase activating protein 4a |
| chr15_+_20416591 | 0.37 |
ENSDART00000161047
|
il15l
|
interleukin 15, like |
| chr22_+_24194444 | 0.36 |
ENSDART00000165380
|
uchl5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr23_-_37010705 | 0.36 |
ENSDART00000134461
|
zgc:193690
|
zgc:193690 |
| chr24_-_21224397 | 0.35 |
ENSDART00000156537
|
spice1
|
spindle and centriole associated protein 1 |
| chr15_+_20416518 | 0.35 |
ENSDART00000161047
|
il15l
|
interleukin 15, like |
| chr13_-_26668719 | 0.34 |
ENSDART00000139264
|
vrk2
|
vaccinia related kinase 2 |
| chr8_-_18480333 | 0.33 |
ENSDART00000149446
|
ENSDARG00000077696
|
ENSDARG00000077696 |
| chr24_+_24581737 | 0.33 |
ENSDART00000137436
|
mtfr1
|
mitochondrial fission regulator 1 |
| chr17_+_51655501 | 0.33 |
ENSDART00000149807
|
odc1
|
ornithine decarboxylase 1 |
| chr8_+_50202152 | 0.33 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr21_-_20908880 | 0.33 |
ENSDART00000079701
|
rnf180
|
ring finger protein 180 |
| chr23_+_33792145 | 0.33 |
ENSDART00000024695
|
dazap2
|
DAZ associated protein 2 |
| chr8_-_37023410 | 0.33 |
ENSDART00000111513
|
si:ch211-218o21.4
|
si:ch211-218o21.4 |
| chr22_+_4145005 | 0.33 |
ENSDART00000171774
|
zgc:171566
|
zgc:171566 |
| chr17_-_43409443 | 0.33 |
ENSDART00000156418
|
itpk1b
|
inositol-tetrakisphosphate 1-kinase b |
| chr9_+_33343936 | 0.32 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr23_-_27318392 | 0.31 |
|
|
|
| chr12_+_46281592 | 0.31 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr4_-_25192290 | 0.31 |
ENSDART00000066930
|
atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr19_-_7124381 | 0.30 |
ENSDART00000104845
|
tapbp.1
|
TAP binding protein (tapasin), tandem duplicate 1 |
| chr12_-_3042545 | 0.30 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr6_-_6818607 | 0.30 |
ENSDART00000151822
|
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr11_-_18074946 | 0.30 |
|
|
|
| chr2_+_32760638 | 0.29 |
ENSDART00000022909
|
klhl18
|
kelch-like family member 18 |
| chr12_-_36113358 | 0.29 |
|
|
|
| chr23_+_4315077 | 0.28 |
ENSDART00000008761
|
arl6ip5b
|
ADP-ribosylation factor-like 6 interacting protein 5b |
| KN150286v1_-_2942 | 0.28 |
|
|
|
| chr6_+_44199738 | 0.28 |
ENSDART00000075486
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr13_+_40347099 | 0.28 |
|
|
|
| chr21_-_30217360 | 0.28 |
ENSDART00000101054
|
dnajc18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr4_+_163079 | 0.28 |
|
|
|
| chr11_-_14044356 | 0.28 |
ENSDART00000085158
|
tmem259
|
transmembrane protein 259 |
| chr12_+_46281511 | 0.27 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr13_-_7904111 | 0.27 |
|
|
|
| chr16_-_16318156 | 0.27 |
ENSDART00000108965
|
gra
|
granulito |
| chr3_+_48810347 | 0.27 |
ENSDART00000133283
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr7_-_25991514 | 0.27 |
ENSDART00000137769
|
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr13_+_12538980 | 0.27 |
ENSDART00000010517
|
eif4eb
|
eukaryotic translation initiation factor 4eb |
| chr7_+_26730804 | 0.26 |
|
|
|
| chr6_+_12810986 | 0.26 |
ENSDART00000104757
ENSDART00000165896 |
casp8
|
caspase 8, apoptosis-related cysteine peptidase |
| chr8_+_50201986 | 0.26 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr1_+_36593187 | 0.25 |
ENSDART00000171340
|
fam193a
|
family with sequence similarity 193, member A |
| chr19_+_10742944 | 0.25 |
ENSDART00000165653
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr15_+_1238560 | 0.25 |
ENSDART00000167760
ENSDART00000163827 |
mfsd1
|
major facilitator superfamily domain containing 1 |
| chr11_-_22210506 | 0.25 |
ENSDART00000146873
|
tmem183a
|
transmembrane protein 183A |
| chr19_-_37533194 | 0.25 |
|
|
|
| chr4_+_1873500 | 0.25 |
ENSDART00000021139
|
fgfr1op2
|
FGFR1 oncogene partner 2 |
| chr8_-_1586037 | 0.24 |
ENSDART00000142703
|
CU694486.1
|
ENSDARG00000091948 |
| chr23_-_24299965 | 0.24 |
ENSDART00000109134
|
plekhm2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr15_+_42279336 | 0.24 |
ENSDART00000114801
|
SGPP2
|
sphingosine-1-phosphate phosphatase 2 |
| chr13_+_15669704 | 0.23 |
ENSDART00000146234
|
apopt1
|
apoptogenic 1, mitochondrial |
| chr17_+_28607337 | 0.23 |
ENSDART00000122260
|
heatr5a
|
HEAT repeat containing 5a |
| chr24_-_33475705 | 0.23 |
ENSDART00000074161
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr9_+_33344121 | 0.23 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr25_+_29752983 | 0.22 |
ENSDART00000141167
|
hsd17b12a
|
hydroxysteroid (17-beta) dehydrogenase 12a |
| chr15_-_1519615 | 0.22 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr6_+_21896033 | 0.22 |
ENSDART00000163394
|
dgke
|
diacylglycerol kinase, epsilon |
| chr17_+_28607470 | 0.21 |
ENSDART00000109693
|
heatr5a
|
HEAT repeat containing 5a |
| chr13_+_20742320 | 0.21 |
|
|
|
| chr23_+_19832770 | 0.20 |
ENSDART00000135820
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr5_-_41523801 | 0.20 |
ENSDART00000051135
|
emc6
|
ER membrane protein complex subunit 6 |
| chr2_-_1028487 | 0.20 |
ENSDART00000058289
|
dusp22b
|
dual specificity phosphatase 22b |
| chr3_-_34687314 | 0.20 |
ENSDART00000084448
|
psmd11a
|
proteasome 26S subunit, non-ATPase 11a |
| chr19_-_7151715 | 0.20 |
ENSDART00000168755
|
tapbp.2
|
TAP binding protein (tapasin), tandem duplicate 2 |
| chr7_+_38408691 | 0.20 |
ENSDART00000171691
|
psmc3
|
proteasome 26S subunit, ATPase 3 |
| chr4_+_76548332 | 0.19 |
ENSDART00000159452
ENSDART00000092415 |
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr7_+_50123055 | 0.19 |
ENSDART00000032324
|
hddc3
|
HD domain containing 3 |
| chr20_+_23602537 | 0.19 |
|
|
|
| chr7_-_71214600 | 0.19 |
ENSDART00000042492
|
sepsecs
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr5_-_3103278 | 0.19 |
|
|
|
| chr15_-_43954596 | 0.19 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr18_+_17736565 | 0.19 |
ENSDART00000090608
|
rspry1
|
ring finger and SPRY domain containing 1 |
| chr7_+_34277682 | 0.18 |
ENSDART00000073447
|
fhod1
|
formin homology 2 domain containing 1 |
| chr24_+_9157328 | 0.18 |
ENSDART00000165780
|
otud1
|
OTU deubiquitinase 1 |
| chr12_-_3042573 | 0.18 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr7_-_68033162 | 0.18 |
|
|
|
| chr16_+_42383980 | 0.17 |
|
|
|
| chr11_-_18439280 | 0.17 |
|
|
|
| chr17_+_51654782 | 0.17 |
ENSDART00000149039
|
odc1
|
ornithine decarboxylase 1 |
| chr8_-_11286377 | 0.16 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr6_+_48349630 | 0.16 |
ENSDART00000064826
|
mov10a
|
Mov10 RISC complex RNA helicase a |
| chr2_+_48090771 | 0.16 |
ENSDART00000135584
|
ftr19
|
finTRIM family, member 19 |
| chr2_+_32760546 | 0.16 |
ENSDART00000022909
|
klhl18
|
kelch-like family member 18 |
| chr22_+_12773735 | 0.16 |
ENSDART00000005720
|
stat1a
|
signal transducer and activator of transcription 1a |
| chr11_-_6975491 | 0.16 |
ENSDART00000129955
|
si:ch211-253b8.2
|
si:ch211-253b8.2 |
| chr7_-_25991876 | 0.16 |
ENSDART00000101109
|
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr16_+_48772036 | 0.16 |
ENSDART00000148709
|
brd2b
|
bromodomain containing 2b |
| chr5_-_41523490 | 0.16 |
ENSDART00000051135
|
emc6
|
ER membrane protein complex subunit 6 |
| chr23_+_24345877 | 0.16 |
ENSDART00000029974
|
clcnk
|
chloride channel K |
| chr3_+_23964015 | 0.15 |
ENSDART00000138270
|
copz2
|
coatomer protein complex, subunit zeta 2 |
| chr6_+_49902765 | 0.15 |
ENSDART00000023515
|
chmp4ba
|
charged multivesicular body protein 4Ba |
| chr15_+_44069851 | 0.15 |
ENSDART00000114352
|
zgc:112998
|
zgc:112998 |
| chr4_-_1952201 | 0.15 |
ENSDART00000135749
|
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr23_-_15234716 | 0.15 |
ENSDART00000158943
|
ndrg3b
|
ndrg family member 3b |
| chr3_-_15959984 | 0.15 |
ENSDART00000140376
|
lasp1
|
LIM and SH3 protein 1 |
| chr22_+_30381331 | 0.15 |
ENSDART00000104751
|
mxi1
|
max interactor 1, dimerization protein |
| chr19_-_42847112 | 0.15 |
ENSDART00000086961
|
vps45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr23_-_27318468 | 0.14 |
|
|
|
| chr23_+_33792005 | 0.14 |
ENSDART00000024695
|
dazap2
|
DAZ associated protein 2 |
| chr13_-_17598311 | 0.14 |
ENSDART00000013011
|
vdac2
|
voltage-dependent anion channel 2 |
| chr5_-_31306608 | 0.14 |
|
|
|
| chr19_+_4208233 | 0.14 |
ENSDART00000172524
|
TRAPPC9
|
trafficking protein particle complex 9 |
| chr11_+_41777220 | 0.14 |
ENSDART00000172846
|
camta1
|
calmodulin binding transcription activator 1 |
| chr10_+_4717721 | 0.14 |
ENSDART00000161789
|
palm2
|
paralemmin 2 |
| chr9_+_23118780 | 0.14 |
ENSDART00000061299
|
tsn
|
translin |
| chr22_+_39012456 | 0.14 |
ENSDART00000176985
|
CABZ01044615.1
|
ENSDARG00000108810 |
| chr9_-_33116897 | 0.14 |
|
|
|
| chr17_+_41313762 | 0.14 |
ENSDART00000128482
|
fosl2
|
fos-like antigen 2 |
| chr21_+_17265099 | 0.13 |
ENSDART00000145057
|
tsc1b
|
tuberous sclerosis 1b |
| chr6_-_2768437 | 0.13 |
|
|
|
| chr19_+_33506239 | 0.13 |
ENSDART00000019459
|
fam91a1
|
family with sequence similarity 91, member A1 |
| chr9_-_33799452 | 0.13 |
ENSDART00000100849
ENSDART00000040843 |
caska
|
calcium/calmodulin-dependent serine protein kinase a |
| chr23_-_35904414 | 0.13 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr21_+_19958025 | 0.13 |
|
|
|
| chr3_-_16056497 | 0.13 |
|
|
|
| chr3_-_39048361 | 0.13 |
ENSDART00000013167
|
retsat
|
retinol saturase (all-trans-retinol 13,14-reductase) |
| chr19_-_44161905 | 0.13 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr21_+_22856217 | 0.12 |
ENSDART00000065565
|
alg8
|
ALG8, alpha-1,3-glucosyltransferase |
| chr5_+_33949469 | 0.12 |
ENSDART00000136787
|
aif1l
|
allograft inflammatory factor 1-like |
| chr4_-_7644310 | 0.12 |
ENSDART00000157751
|
BX649411.1
|
ENSDARG00000101254 |
| KN150158v1_-_15806 | 0.12 |
ENSDART00000163880
|
CABZ01067681.1
|
ENSDARG00000103190 |
| chr7_-_30654776 | 0.12 |
ENSDART00000075421
|
sord
|
sorbitol dehydrogenase |
| chr4_-_1952230 | 0.12 |
ENSDART00000135749
|
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr11_+_44738699 | 0.12 |
ENSDART00000172301
ENSDART00000170118 |
tk1
|
thymidine kinase 1, soluble |
| chr8_-_11286437 | 0.12 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr5_+_33949689 | 0.12 |
ENSDART00000139317
|
aif1l
|
allograft inflammatory factor 1-like |
| chr7_+_44366144 | 0.12 |
ENSDART00000163821
|
BX511128.2
|
ENSDARG00000101493 |
| chr15_+_20416637 | 0.12 |
ENSDART00000161047
|
il15l
|
interleukin 15, like |
| chr8_-_18480580 | 0.11 |
ENSDART00000149446
|
ENSDARG00000077696
|
ENSDARG00000077696 |
| chr8_+_48976657 | 0.11 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr8_-_11286308 | 0.11 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr23_-_20084226 | 0.11 |
ENSDART00000163396
|
b4galt3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr8_-_18637019 | 0.11 |
ENSDART00000172584
ENSDART00000100516 |
stap2b
|
signal transducing adaptor family member 2b |
| chr3_+_32802124 | 0.11 |
ENSDART00000112742
|
nbr1b
|
neighbor of brca1 gene 1b |
| chr17_-_31027949 | 0.11 |
ENSDART00000152016
|
eml1
|
echinoderm microtubule associated protein like 1 |
| chr5_+_60823598 | 0.11 |
ENSDART00000023676
|
bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr22_+_17803347 | 0.10 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr22_+_39110524 | 0.10 |
ENSDART00000153841
|
lmcd1
|
LIM and cysteine-rich domains 1 |
| chr15_-_809395 | 0.10 |
|
|
|
| KN150711v1_-_5452 | 0.10 |
|
|
|
| chr23_+_25808393 | 0.10 |
|
|
|
| chr13_+_5958225 | 0.10 |
ENSDART00000049328
|
fam120b
|
family with sequence similarity 120B |
| chr18_+_7584201 | 0.10 |
ENSDART00000163709
|
zgc:77650
|
zgc:77650 |
| chr7_+_20829815 | 0.10 |
ENSDART00000173612
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr18_+_15873032 | 0.10 |
ENSDART00000141800
|
eea1
|
early endosome antigen 1 |
| chr1_-_51863300 | 0.10 |
ENSDART00000004233
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr7_-_6332802 | 0.09 |
ENSDART00000147978
|
HIST2H2AB (1 of many)
|
histone cluster 2 H2A family member b |
| chr3_-_32231069 | 0.09 |
ENSDART00000035545
|
prmt1
|
protein arginine methyltransferase 1 |
| KN150359v1_+_69830 | 0.09 |
|
|
|
| chr23_-_37010664 | 0.09 |
ENSDART00000102886
|
zgc:193690
|
zgc:193690 |
| chr13_+_12539027 | 0.09 |
ENSDART00000010517
|
eif4eb
|
eukaryotic translation initiation factor 4eb |
| chr19_-_7151801 | 0.09 |
ENSDART00000176808
|
tapbp.1
|
TAP binding protein (tapasin), tandem duplicate 1 |
| chr19_-_41820114 | 0.09 |
ENSDART00000038038
|
shfm1
|
split hand/foot malformation (ectrodactyly) type 1 |
| chr9_+_31204464 | 0.09 |
|
|
|
| chr6_-_7625286 | 0.09 |
|
|
|
| chr2_+_52529869 | 0.09 |
ENSDART00000111266
|
theg
|
theg spermatid protein |
| chr1_+_9633924 | 0.09 |
ENSDART00000029774
|
tmem55bb
|
transmembrane protein 55Bb |
| chr5_+_60823823 | 0.09 |
ENSDART00000023676
|
bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr16_-_13927947 | 0.08 |
ENSDART00000144856
|
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr3_-_30117381 | 0.08 |
ENSDART00000077029
|
emc10
|
ER membrane protein complex subunit 10 |
| chr16_-_13927684 | 0.08 |
ENSDART00000146719
|
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr5_-_13844114 | 0.08 |
|
|
|
| chr13_+_28655188 | 0.08 |
ENSDART00000039028
|
nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chr9_-_30692456 | 0.08 |
|
|
|
| chr4_-_17271996 | 0.07 |
ENSDART00000145513
|
lrmp
|
lymphoid-restricted membrane protein |
| chr4_-_18786577 | 0.07 |
ENSDART00000141187
|
def6c
|
differentially expressed in FDCP 6c homolog |
| chr7_-_71214683 | 0.07 |
ENSDART00000042492
|
sepsecs
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr10_-_28474052 | 0.07 |
|
|
|
| chr4_-_4526152 | 0.07 |
ENSDART00000057519
|
zgc:194209
|
zgc:194209 |
| chr23_-_4769182 | 0.07 |
ENSDART00000144536
|
cnbpa
|
CCHC-type zinc finger, nucleic acid binding protein a |
| chr4_+_25192397 | 0.07 |
ENSDART00000078529
ENSDART00000136643 |
kin
|
Kin17 DNA and RNA binding protein |
| chr12_-_27121226 | 0.07 |
ENSDART00000153101
|
psme3
|
proteasome activator subunit 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.2 | 0.6 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.1 | 0.8 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.1 | 1.1 | GO:0035283 | fourth ventricle development(GO:0021592) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.5 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.4 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 0.3 | GO:0043903 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) interaction with host(GO:0051701) |
| 0.1 | 0.4 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.3 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.8 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 0.2 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.4 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.1 | 0.3 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 0.4 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.2 | GO:0090148 | membrane fission(GO:0090148) |
| 0.1 | 0.2 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.3 | GO:0009712 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.0 | 0.1 | GO:1905207 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.0 | 0.4 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.3 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.0 | 0.3 | GO:1901908 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.4 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.5 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.2 | GO:0008210 | estrogen biosynthetic process(GO:0006703) estrogen metabolic process(GO:0008210) |
| 0.0 | 0.1 | GO:0007344 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.0 | 0.7 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 0.0 | 0.2 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.2 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.3 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.1 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.2 | GO:0060337 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.3 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.3 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.1 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.4 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.2 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 1.0 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.6 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.5 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.1 | 0.4 | GO:0052725 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.1 | 0.4 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 1.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:0090624 | endoribonuclease activity, cleaving miRNA-paired mRNA(GO:0090624) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.4 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.3 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.3 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0016843 | amine-lyase activity(GO:0016843) strictosidine synthase activity(GO:0016844) |
| 0.0 | 0.1 | GO:0016504 | peptidase activator activity(GO:0016504) endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.8 | GO:0005126 | cytokine receptor binding(GO:0005126) |
| 0.0 | 0.4 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.5 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |