Project
DANIO-CODE
Navigation
Downloads
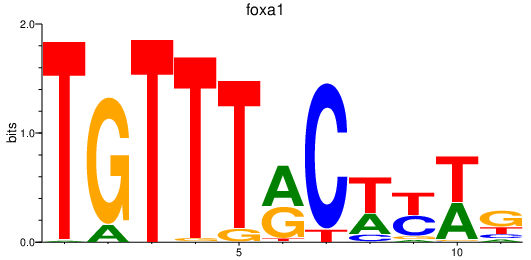
Results for foxa1_foxa_foxa2
Z-value: 2.11
Transcription factors associated with foxa1_foxa_foxa2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxa1
|
ENSDARG00000102138 | forkhead box A1 |
|
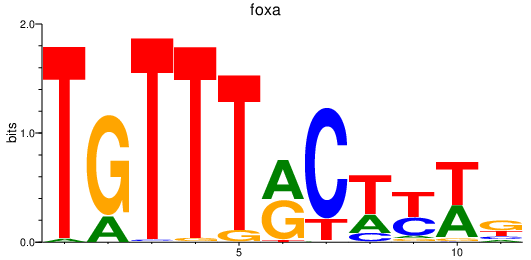
foxa
|
ENSDARG00000087094 | forkhead box A sequence |
|
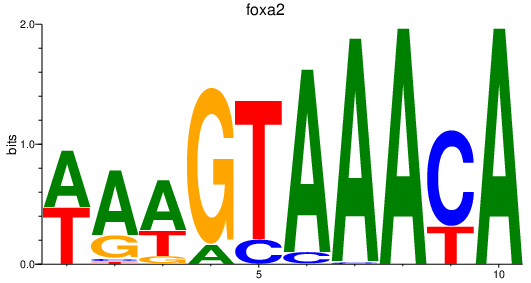
foxa2
|
ENSDARG00000003411 | forkhead box A2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxa | dr10_dc_chr14_-_33641145_33641206 | -0.92 | 4.5e-07 | Click! |
| foxa1 | dr10_dc_chr17_+_10161999_10162052 | 0.92 | 5.4e-07 | Click! |
| foxa2 | dr10_dc_chr17_-_42293039_42293129 | 0.67 | 4.8e-03 | Click! |
Activity profile of foxa1_foxa_foxa2 motif
Sorted Z-values of foxa1_foxa_foxa2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of foxa1_foxa_foxa2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_31501522 | 10.57 |
ENSDART00000144422
|
cpn1
|
carboxypeptidase N, polypeptide 1 |
| chr23_+_32573474 | 8.21 |
ENSDART00000134811
|
si:dkey-261h17.1
|
si:dkey-261h17.1 |
| chr9_+_54185607 | 7.29 |
ENSDART00000165991
|
lect1
|
leukocyte cell derived chemotaxin 1 |
| chr5_+_49093250 | 6.68 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr16_+_24063849 | 6.41 |
ENSDART00000135084
|
apoa2
|
apolipoprotein A-II |
| chr5_+_49093134 | 6.21 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr16_+_5466342 | 6.17 |
ENSDART00000160008
|
plecb
|
plectin b |
| chr14_-_25879853 | 6.00 |
ENSDART00000014149
|
fat2
|
FAT atypical cadherin 2 |
| chr4_+_17428131 | 5.97 |
ENSDART00000056005
|
ascl1a
|
achaete-scute family bHLH transcription factor 1a |
| chr13_+_1377875 | 5.92 |
|
|
|
| chr15_+_37039861 | 5.92 |
ENSDART00000172664
|
kirrel3l
|
kin of IRRE like 3 like |
| chr5_+_37053530 | 5.90 |
ENSDART00000161051
|
sptbn2
|
spectrin, beta, non-erythrocytic 2 |
| chr25_+_33527870 | 5.76 |
ENSDART00000011967
|
anxa2a
|
annexin A2a |
| chr6_-_32718634 | 5.43 |
ENSDART00000175666
|
mafaa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Aa |
| chr23_+_22977031 | 5.22 |
|
|
|
| chr22_-_36915567 | 4.98 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr23_+_33362843 | 4.97 |
|
|
|
| chr6_-_14821305 | 4.90 |
|
|
|
| chr4_+_10722966 | 4.79 |
ENSDART00000102534
ENSDART00000067253 ENSDART00000136000 |
stab2
|
stabilin 2 |
| chr2_+_38178934 | 4.57 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr7_+_48532749 | 4.41 |
ENSDART00000145375
|
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr11_-_29316162 | 4.17 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr3_+_56970554 | 3.94 |
ENSDART00000162930
|
bahcc1a
|
BAH domain and coiled-coil containing 1a |
| chr13_+_16130212 | 3.93 |
ENSDART00000125813
|
mocos
|
molybdenum cofactor sulfurase |
| chr4_-_13996902 | 3.92 |
ENSDART00000147955
|
prickle1b
|
prickle homolog 1b |
| chr9_-_46272322 | 3.78 |
ENSDART00000169682
|
hdac4
|
histone deacetylase 4 |
| chr9_+_52872108 | 3.77 |
ENSDART00000157453
ENSDART00000163684 |
nme8
|
NME/NM23 family member 8 |
| chr7_-_52283383 | 3.74 |
ENSDART00000165649
|
tcf12
|
transcription factor 12 |
| chr4_-_73485204 | 3.69 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| chr16_-_54634679 | 3.66 |
ENSDART00000075275
ENSDART00000115024 |
pklr
|
pyruvate kinase, liver and RBC |
| chr17_-_21260319 | 3.66 |
ENSDART00000166524
|
hspa12a
|
heat shock protein 12A |
| chr17_+_24018420 | 3.65 |
ENSDART00000140767
|
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr10_+_32007448 | 3.62 |
ENSDART00000019416
|
lhfp
|
lipoma HMGIC fusion partner |
| chr24_+_20430778 | 3.57 |
ENSDART00000010488
|
klhl40b
|
kelch-like family member 40b |
| chr13_+_48324283 | 3.57 |
ENSDART00000177067
|
CU929120.1
|
ENSDARG00000107321 |
| chr21_-_37286942 | 3.56 |
ENSDART00000100310
|
dbn1
|
drebrin 1 |
| chr8_+_49582361 | 3.51 |
ENSDART00000108613
|
rasef
|
RAS and EF-hand domain containing |
| chr6_-_40660116 | 3.48 |
ENSDART00000154359
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr20_-_22576513 | 3.45 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr13_+_1595986 | 3.33 |
|
|
|
| KN150702v1_+_177145 | 3.32 |
ENSDART00000166225
|
CABZ01076674.1
|
ENSDARG00000102620 |
| chr23_+_11412329 | 3.29 |
ENSDART00000135406
|
chl1a
|
cell adhesion molecule L1-like a |
| chr18_+_40364675 | 3.23 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr17_+_25500291 | 3.22 |
ENSDART00000041721
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr9_-_19154264 | 3.18 |
ENSDART00000081878
|
pou1f1
|
POU class 1 homeobox 1 |
| chr23_+_35819625 | 3.05 |
ENSDART00000049551
|
rarga
|
retinoic acid receptor gamma a |
| chr13_-_31516399 | 3.02 |
ENSDART00000172375
ENSDART00000125987 ENSDART00000143903 ENSDART00000158719 |
six4a
|
SIX homeobox 4a |
| chr20_-_20634087 | 3.01 |
ENSDART00000125039
|
six6b
|
SIX homeobox 6b |
| chr15_+_7179382 | 3.00 |
ENSDART00000101578
|
her8.2
|
hairy-related 8.2 |
| chr11_-_4216204 | 2.97 |
ENSDART00000121716
|
ENSDARG00000086300
|
ENSDARG00000086300 |
| chr4_-_13922285 | 2.90 |
ENSDART00000080334
|
yaf2
|
YY1 associated factor 2 |
| chr4_-_16835283 | 2.88 |
ENSDART00000165289
|
gys2
|
glycogen synthase 2 |
| chr6_+_24717985 | 2.87 |
ENSDART00000165609
|
barhl2
|
BarH-like homeobox 2 |
| chr12_-_30988279 | 2.87 |
ENSDART00000122972
ENSDART00000005562 ENSDART00000031408 ENSDART00000125046 ENSDART00000009237 |
tcf7l2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr6_-_43679553 | 2.82 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr6_-_16156494 | 2.80 |
|
|
|
| chr6_-_30876091 | 2.79 |
ENSDART00000155330
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr11_-_29910947 | 2.79 |
ENSDART00000156121
|
scml2
|
sex comb on midleg-like 2 (Drosophila) |
| chr21_-_11539798 | 2.77 |
ENSDART00000144770
|
cast
|
calpastatin |
| chr1_-_18695214 | 2.76 |
|
|
|
| chr23_-_18203680 | 2.72 |
ENSDART00000016976
|
nucks1b
|
nuclear casein kinase and cyclin-dependent kinase substrate 1b |
| chr10_+_35535209 | 2.70 |
ENSDART00000109705
|
phldb2a
|
pleckstrin homology-like domain, family B, member 2a |
| chr7_+_48532543 | 2.68 |
ENSDART00000145375
|
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr22_-_36915720 | 2.67 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr11_-_44724371 | 2.62 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr21_-_11561855 | 2.57 |
ENSDART00000162426
|
cast
|
calpastatin |
| chr2_+_5651428 | 2.57 |
ENSDART00000111220
|
mb21d2
|
Mab-21 domain containing 2 |
| chr9_-_39195468 | 2.56 |
ENSDART00000014207
|
myl1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr2_+_17112049 | 2.55 |
ENSDART00000125647
|
tfa
|
transferrin-a |
| chr6_-_31160132 | 2.53 |
ENSDART00000153592
|
CR847887.1
|
ENSDARG00000097226 |
| chr1_+_41148042 | 2.52 |
ENSDART00000145170
ENSDART00000136879 |
smox
|
spermine oxidase |
| chr15_+_24708696 | 2.50 |
ENSDART00000043292
|
smtnl
|
smoothelin, like |
| chr16_-_42011213 | 2.48 |
ENSDART00000111956
|
gramd1a
|
GRAM domain containing 1A |
| chr17_+_32619043 | 2.47 |
ENSDART00000087565
|
eva1a
|
eva-1 homolog A (C. elegans) |
| chr7_+_69119748 | 2.43 |
ENSDART00000177049
|
ENSDARG00000106989
|
ENSDARG00000106989 |
| chr7_+_36627685 | 2.41 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| KN150266v1_+_74105 | 2.41 |
|
|
|
| chr14_+_16632329 | 2.41 |
ENSDART00000163013
|
limch1b
|
LIM and calponin homology domains 1b |
| chr8_-_14146621 | 2.40 |
ENSDART00000063817
|
ndufb11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11 |
| chr13_-_49695322 | 2.39 |
|
|
|
| chr10_+_6316944 | 2.37 |
ENSDART00000162428
|
tpm2
|
tropomyosin 2 (beta) |
| chr21_+_20734431 | 2.36 |
ENSDART00000079732
|
oxct1b
|
3-oxoacid CoA transferase 1b |
| chr5_-_25509259 | 2.33 |
ENSDART00000137376
|
BX510923.1
|
ENSDARG00000094488 |
| chr7_+_17740815 | 2.31 |
ENSDART00000173679
|
ehbp1l1a
|
EH domain binding protein 1-like 1a |
| chr4_+_7668939 | 2.27 |
ENSDART00000149218
|
elk3
|
ELK3, ETS-domain protein |
| chr16_-_2972770 | 2.25 |
ENSDART00000082522
|
abcd2
|
ATP-binding cassette, sub-family D (ALD), member 2 |
| chr3_-_21149752 | 2.24 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr24_-_16994956 | 2.18 |
ENSDART00000111079
|
CR376737.1
|
ENSDARG00000077318 |
| chr1_-_24486146 | 2.18 |
ENSDART00000144711
|
tmem154
|
transmembrane protein 154 |
| chr3_-_49823285 | 2.16 |
ENSDART00000169771
|
pmp22a
|
peripheral myelin protein 22a |
| chr16_-_21981065 | 2.16 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr13_+_30546398 | 2.15 |
|
|
|
| chr10_+_44364117 | 2.15 |
|
|
|
| chr25_+_4490129 | 2.13 |
|
|
|
| chr24_+_13780640 | 2.12 |
ENSDART00000136443
ENSDART00000081595 ENSDART00000012253 |
eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr24_-_33617592 | 2.10 |
|
|
|
| chr5_-_63422783 | 2.09 |
ENSDART00000083684
|
pappab
|
pregnancy-associated plasma protein A, pappalysin 1b |
| chr14_-_6624878 | 2.08 |
ENSDART00000166439
|
si:ch211-266k2.1
|
si:ch211-266k2.1 |
| chr19_-_35739239 | 2.05 |
|
|
|
| chr7_+_19858190 | 2.05 |
ENSDART00000179395
ENSDART00000113793 ENSDART00000169603 |
zgc:114045
|
zgc:114045 |
| chr14_+_33382973 | 2.02 |
ENSDART00000132488
|
apln
|
apelin |
| chr4_+_25296408 | 1.96 |
ENSDART00000174536
|
sfmbt2
|
Scm-like with four mbt domains 2 |
| chr6_-_8501230 | 1.96 |
ENSDART00000143956
|
cavin2b
|
caveolae associated protein 2b |
| chr8_-_11806627 | 1.96 |
ENSDART00000064017
|
rapgef1a
|
Rap guanine nucleotide exchange factor (GEF) 1a |
| chr5_-_25509188 | 1.94 |
ENSDART00000137376
|
BX510923.1
|
ENSDARG00000094488 |
| chr5_-_62187774 | 1.94 |
|
|
|
| chr11_-_15162109 | 1.92 |
ENSDART00000124968
|
rpn2
|
ribophorin II |
| chr18_-_16806665 | 1.92 |
ENSDART00000048722
|
ampd3b
|
adenosine monophosphate deaminase 3b |
| chr21_-_14534576 | 1.86 |
ENSDART00000089967
ENSDART00000132142 |
cacna1bb
|
calcium channel, voltage-dependent, N type, alpha 1B subunit, b |
| chr11_+_41324286 | 1.85 |
ENSDART00000055710
|
aldh4a1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr12_-_8466427 | 1.85 |
ENSDART00000135865
|
egr2b
|
early growth response 2b |
| chr5_-_25509310 | 1.85 |
ENSDART00000137376
|
BX510923.1
|
ENSDARG00000094488 |
| chr3_-_34208423 | 1.85 |
ENSDART00000151634
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr10_-_17274395 | 1.85 |
ENSDART00000135679
|
rab36
|
RAB36, member RAS oncogene family |
| chr11_+_6809190 | 1.84 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr1_+_35058654 | 1.84 |
ENSDART00000131284
|
hhip
|
hedgehog interacting protein |
| chr12_+_6065116 | 1.84 |
ENSDART00000139054
|
sgms1
|
sphingomyelin synthase 1 |
| chr4_-_73485028 | 1.82 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| chr13_+_27086152 | 1.82 |
ENSDART00000172079
|
BX323884.1
|
ENSDARG00000091897 |
| chr4_+_25296376 | 1.82 |
ENSDART00000174536
|
sfmbt2
|
Scm-like with four mbt domains 2 |
| chr13_+_16130123 | 1.79 |
ENSDART00000170855
|
mocos
|
molybdenum cofactor sulfurase |
| chr14_-_30467807 | 1.79 |
ENSDART00000173262
|
prss23
|
protease, serine, 23 |
| chr9_+_54185729 | 1.78 |
ENSDART00000165991
|
lect1
|
leukocyte cell derived chemotaxin 1 |
| chr16_-_29342511 | 1.77 |
ENSDART00000058870
|
rhbg
|
Rh family, B glycoprotein (gene/pseudogene) |
| chr24_+_21200975 | 1.77 |
ENSDART00000126519
|
shisa2b
|
shisa family member 2b |
| chr19_+_16128021 | 1.77 |
ENSDART00000046530
|
rab42a
|
RAB42, member RAS oncogene family a |
| chr23_+_28843683 | 1.75 |
ENSDART00000132179
|
masp2
|
mannan-binding lectin serine peptidase 2 |
| chr7_-_58427369 | 1.75 |
ENSDART00000149347
|
chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr16_-_30606364 | 1.73 |
ENSDART00000147986
|
lmna
|
lamin A |
| chr16_-_31670211 | 1.71 |
ENSDART00000138216
|
CR855311.1
|
ENSDARG00000090352 |
| chr9_-_24219908 | 1.70 |
ENSDART00000144163
|
rpe
|
ribulose-5-phosphate-3-epimerase |
| chr23_-_27226280 | 1.69 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr14_-_33704021 | 1.68 |
ENSDART00000149396
ENSDART00000123607 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr12_+_22282872 | 1.66 |
ENSDART00000153055
|
hdlbpb
|
high density lipoprotein binding protein b |
| chr20_+_23392782 | 1.66 |
|
|
|
| chr10_+_39148251 | 1.66 |
ENSDART00000125986
|
ENSDARG00000055339
|
ENSDARG00000055339 |
| chr4_-_17735989 | 1.65 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr4_-_13568659 | 1.64 |
ENSDART00000169582
|
mdm1
|
Mdm1 nuclear protein homolog (mouse) |
| chr22_-_14103803 | 1.64 |
ENSDART00000062902
|
ENSDARG00000042857
|
ENSDARG00000042857 |
| chr5_+_34022151 | 1.61 |
ENSDART00000141338
|
enc1
|
ectodermal-neural cortex 1 |
| chr6_+_14822990 | 1.59 |
ENSDART00000149949
|
pou3f3b
|
POU class 3 homeobox 3b |
| chr14_+_38375351 | 1.58 |
ENSDART00000175744
|
BX088707.3
|
ENSDARG00000107351 |
| chr23_-_30772173 | 1.58 |
ENSDART00000138841
|
CR456642.1
|
ENSDARG00000092307 |
| chr5_+_29327363 | 1.57 |
|
|
|
| chr21_-_22914947 | 1.55 |
ENSDART00000083449
|
dub
|
duboraya |
| chr6_+_28886741 | 1.54 |
ENSDART00000065137
|
tp63
|
tumor protein p63 |
| chr1_-_22144014 | 1.54 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr20_+_25326042 | 1.54 |
ENSDART00000130494
|
moxd1
|
monooxygenase, DBH-like 1 |
| chr18_+_26916897 | 1.54 |
ENSDART00000050230
|
tspan3a
|
tetraspanin 3a |
| chr17_+_27417635 | 1.52 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr23_-_30861356 | 1.51 |
ENSDART00000114628
|
myt1a
|
myelin transcription factor 1a |
| chr4_-_13568618 | 1.51 |
ENSDART00000169582
|
mdm1
|
Mdm1 nuclear protein homolog (mouse) |
| chr15_+_37040064 | 1.49 |
ENSDART00000172664
|
kirrel3l
|
kin of IRRE like 3 like |
| chr8_-_23102555 | 1.48 |
|
|
|
| chr19_-_29205158 | 1.47 |
ENSDART00000026992
|
sox4a
|
SRY (sex determining region Y)-box 4a |
| chr6_+_14823185 | 1.47 |
ENSDART00000149202
|
pou3f3b
|
POU class 3 homeobox 3b |
| chr12_-_48278273 | 1.46 |
|
|
|
| chr23_+_36016450 | 1.45 |
ENSDART00000103035
|
hoxc6a
|
homeobox C6a |
| chr20_-_3970778 | 1.45 |
ENSDART00000178724
ENSDART00000178565 |
TRIM67
|
tripartite motif containing 67 |
| chr4_-_18660286 | 1.45 |
ENSDART00000178638
|
pparaa
|
peroxisome proliferator-activated receptor alpha a |
| chr10_-_10372266 | 1.45 |
|
|
|
| chr15_+_36329506 | 1.43 |
ENSDART00000099501
|
masp1
|
mannan-binding lectin serine peptidase 1 |
| chr3_-_42649955 | 1.40 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr6_+_10491884 | 1.40 |
ENSDART00000151017
|
BX936397.2
|
ENSDARG00000096373 |
| chr5_+_29327479 | 1.39 |
|
|
|
| chr25_-_34855094 | 1.39 |
ENSDART00000171917
|
SVIP
|
small VCP interacting protein |
| chr7_-_57796486 | 1.38 |
ENSDART00000043984
|
ank2b
|
ankyrin 2b, neuronal |
| chr24_-_4941954 | 1.37 |
ENSDART00000127597
|
zic1
|
zic family member 1 (odd-paired homolog, Drosophila) |
| chr25_+_23326562 | 1.37 |
|
|
|
| chr12_-_13042845 | 1.35 |
ENSDART00000152510
|
ccser2b
|
coiled-coil serine-rich protein 2b |
| chr22_-_14103942 | 1.35 |
ENSDART00000140323
|
ENSDARG00000042857
|
ENSDARG00000042857 |
| chr25_-_14953284 | 1.34 |
ENSDART00000165632
|
pax6a
|
paired box 6a |
| chr8_+_26799298 | 1.34 |
ENSDART00000134987
|
si:ch211-156j16.1
|
si:ch211-156j16.1 |
| chr3_-_8608620 | 1.34 |
|
|
|
| chr10_+_26786051 | 1.32 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr24_-_25545773 | 1.30 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr15_-_25430003 | 1.30 |
ENSDART00000152651
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr8_+_23763721 | 1.28 |
ENSDART00000062968
|
si:ch211-163l21.8
|
si:ch211-163l21.8 |
| chr12_-_304698 | 1.26 |
|
|
|
| chr10_+_39148457 | 1.25 |
ENSDART00000125986
|
ENSDARG00000055339
|
ENSDARG00000055339 |
| chr12_-_2487745 | 1.23 |
ENSDART00000022471
ENSDART00000145213 |
mapk8b
|
mitogen-activated protein kinase 8b |
| chr17_-_26849495 | 1.22 |
ENSDART00000153590
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr1_+_50395721 | 1.22 |
ENSDART00000134065
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr10_-_36690119 | 1.21 |
ENSDART00000077161
|
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 |
| chr23_-_27226387 | 1.20 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr13_-_37339615 | 1.19 |
ENSDART00000100324
|
ppp2r5eb
|
protein phosphatase 2, regulatory subunit B', epsilon isoform b |
| chr21_+_11410738 | 1.18 |
ENSDART00000146701
|
si:dkey-184p9.7
|
si:dkey-184p9.7 |
| chr24_-_21353742 | 1.18 |
ENSDART00000153695
|
atp8a2
|
ATPase, aminophospholipid transporter, class I, type 8A, member 2 |
| chr13_-_30515486 | 1.17 |
ENSDART00000139607
|
zcchc24
|
zinc finger, CCHC domain containing 24 |
| chr13_-_40628582 | 1.16 |
ENSDART00000111641
ENSDART00000159255 |
morn4
|
MORN repeat containing 4 |
| chr20_+_6600246 | 1.16 |
ENSDART00000136567
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr13_+_28997608 | 1.15 |
ENSDART00000109546
|
unc5b
|
unc-5 netrin receptor B |
| chr22_-_36915689 | 1.14 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr9_+_21911860 | 1.14 |
ENSDART00000102021
|
sox1a
|
SRY (sex determining region Y)-box 1a |
| chr8_+_50498398 | 1.14 |
|
|
|
| chr1_-_10423289 | 1.12 |
|
|
|
| chr23_-_20332076 | 1.12 |
ENSDART00000147326
|
lamb2
|
laminin, beta 2 (laminin S) |
| chr20_+_29307142 | 1.08 |
ENSDART00000153016
|
katnbl1
|
katanin p80 subunit B-like 1 |
| KN150520v1_+_33937 | 1.07 |
|
|
|
| chr4_+_1680973 | 1.07 |
ENSDART00000149826
ENSDART00000166437 ENSDART00000031135 ENSDART00000172300 ENSDART00000067446 |
slc38a4
|
solute carrier family 38, member 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.0 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 1.3 | 3.8 | GO:0036073 | perichondral ossification(GO:0036073) |
| 1.3 | 8.8 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 1.2 | 4.8 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) positive regulation of lymphangiogenesis(GO:1901492) |
| 1.2 | 3.6 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 1.2 | 7.1 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 1.2 | 12.9 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 1.0 | 2.9 | GO:0042756 | drinking behavior(GO:0042756) |
| 1.0 | 5.7 | GO:0043545 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.9 | 4.6 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.9 | 3.6 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.8 | 2.5 | GO:0019730 | antimicrobial humoral response(GO:0019730) antibacterial humoral response(GO:0019731) |
| 0.8 | 2.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.7 | 2.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.7 | 2.9 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.7 | 6.3 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.6 | 3.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.6 | 1.8 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.6 | 2.4 | GO:0046950 | cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.6 | 1.7 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.6 | 1.7 | GO:0042423 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.5 | 2.9 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.5 | 10.6 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.5 | 4.8 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.5 | 6.6 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.5 | 1.8 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.5 | 1.8 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.4 | 7.4 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.4 | 1.6 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.4 | 6.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.4 | 3.4 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.4 | 2.3 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.3 | 2.8 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.3 | 1.4 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.3 | 1.9 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.3 | 0.9 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.3 | 1.5 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.3 | 1.8 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.3 | 2.5 | GO:0006956 | complement activation(GO:0006956) |
| 0.3 | 0.8 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.3 | 3.7 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.3 | 0.8 | GO:0033280 | response to vitamin D(GO:0033280) |
| 0.3 | 1.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 1.7 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.2 | 1.7 | GO:0001964 | startle response(GO:0001964) |
| 0.2 | 1.8 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 1.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.2 | 0.9 | GO:0008206 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.2 | 1.9 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 1.4 | GO:0010888 | negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) |
| 0.2 | 1.9 | GO:0048931 | lateral line nerve glial cell differentiation(GO:0048895) myelination of lateral line nerve axons(GO:0048897) posterior lateral line nerve glial cell differentiation(GO:0048931) myelination of posterior lateral line nerve axons(GO:0048932) lateral line nerve glial cell development(GO:0048937) lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048938) posterior lateral line nerve glial cell development(GO:0048941) posterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048942) |
| 0.2 | 0.8 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.2 | 2.0 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.2 | 2.9 | GO:0009743 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.2 | 1.8 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.2 | 0.5 | GO:0032675 | interleukin-6 production(GO:0032635) regulation of interleukin-6 production(GO:0032675) |
| 0.2 | 0.5 | GO:0014005 | microglia development(GO:0014005) |
| 0.1 | 0.9 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.1 | 1.5 | GO:0003306 | Wnt signaling pathway involved in heart development(GO:0003306) |
| 0.1 | 0.4 | GO:0055091 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) reverse cholesterol transport(GO:0043691) phospholipid homeostasis(GO:0055091) |
| 0.1 | 5.2 | GO:0051693 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) actin filament capping(GO:0051693) |
| 0.1 | 3.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 6.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 0.3 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.1 | 1.2 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.1 | 0.4 | GO:0035022 | positive regulation of cell-matrix adhesion(GO:0001954) positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 1.1 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.0 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.1 | 0.6 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.1 | 0.4 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.1 | 0.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 3.2 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 0.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.5 | GO:1900153 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.1 | 0.7 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 1.5 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 1.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.4 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.1 | 6.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.0 | 2.7 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.5 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.9 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.5 | GO:0000272 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.3 | GO:0007596 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.0 | 1.9 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.0 | 0.7 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.6 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 2.4 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 3.4 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 1.0 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.3 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.2 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 1.0 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.7 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.8 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 1.8 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 2.9 | GO:0001708 | cell fate specification(GO:0001708) |
| 0.0 | 0.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 1.2 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.4 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.7 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 4.5 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 0.6 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.4 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.2 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 1.2 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 2.4 | GO:0043066 | negative regulation of apoptotic process(GO:0043066) |
| 0.0 | 1.8 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 3.5 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 1.1 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 0.3 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 3.9 | GO:0008283 | cell proliferation(GO:0008283) |
| 0.0 | 5.1 | GO:0022610 | cell adhesion(GO:0007155) biological adhesion(GO:0022610) |
| 0.0 | 0.4 | GO:0045165 | cell fate commitment(GO:0045165) |
| 0.0 | 0.7 | GO:0060538 | skeletal muscle organ development(GO:0060538) |
| 0.0 | 0.1 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 1.6 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 3.7 | GO:0043009 | chordate embryonic development(GO:0043009) |
| 0.0 | 0.5 | GO:0099643 | neurotransmitter secretion(GO:0007269) presynaptic process involved in chemical synaptic transmission(GO:0099531) signal release from synapse(GO:0099643) |
| 0.0 | 1.0 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 3.3 | GO:0007507 | heart development(GO:0007507) |
| 0.0 | 0.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.9 | GO:0008091 | spectrin(GO:0008091) |
| 0.7 | 2.9 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.6 | 2.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.6 | 6.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.4 | 1.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.2 | 3.9 | GO:0031672 | A band(GO:0031672) |
| 0.2 | 6.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.2 | 2.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 1.9 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 0.9 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.2 | 1.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 0.8 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 2.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.3 | GO:0098594 | mucin granule(GO:0098594) |
| 0.1 | 1.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.5 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.1 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 3.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 3.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 5.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 1.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 1.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 3.0 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 0.4 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 8.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 7.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.1 | 12.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 2.4 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 2.0 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 3.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 29.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.6 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 5.7 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 3.8 | GO:0005912 | adherens junction(GO:0005912) anchoring junction(GO:0070161) |
| 0.0 | 1.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 2.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 7.6 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 4.1 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 3.8 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 4.1 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.8 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.6 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.0 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.4 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.9 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.0 | 3.1 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.7 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 1.2 | 3.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 1.0 | 7.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.9 | 10.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.7 | 5.8 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.7 | 2.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.7 | 6.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.6 | 2.4 | GO:0032028 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.6 | 2.4 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.6 | 6.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.5 | 1.6 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.5 | 2.9 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.4 | 2.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 1.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.4 | 1.5 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.4 | 1.9 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 4.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 3.8 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.3 | 1.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 1.8 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.3 | 8.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.3 | 0.8 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.3 | 2.9 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.3 | 3.7 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.3 | 18.4 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.2 | 2.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 1.8 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.2 | 1.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 0.5 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.2 | 0.9 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.2 | 2.3 | GO:0015245 | long-chain fatty acid transporter activity(GO:0005324) fatty acid transporter activity(GO:0015245) |
| 0.2 | 2.4 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.2 | 5.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.7 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 0.5 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 1.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.6 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 1.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 2.3 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 1.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.5 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 3.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 1.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.4 | GO:0016917 | G-protein coupled GABA receptor activity(GO:0004965) GABA receptor activity(GO:0016917) |
| 0.1 | 2.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 5.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 8.5 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.1 | 3.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 5.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 1.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 3.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 3.4 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 6.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 2.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.0 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 1.6 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 3.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 2.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 6.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 33.4 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.4 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.4 | GO:0031210 | phosphatidylcholine binding(GO:0031210) quaternary ammonium group binding(GO:0050997) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 3.9 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.7 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 1.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.8 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 3.8 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 8.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.4 | 3.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 1.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 3.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 3.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.8 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 1.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 3.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 0.7 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.1 | 1.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 3.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 2.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 1.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 3.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 8.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.8 | 5.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.8 | 6.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.8 | 3.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.8 | 2.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.7 | 4.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 2.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 4.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 3.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 2.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 1.3 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.2 | 0.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.1 | 3.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 5.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 3.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 2.9 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.1 | 1.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.4 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.1 | 0.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 3.3 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.8 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 1.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 1.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.6 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 2.3 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 1.7 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 0.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |