Project
DANIO-CODE
Navigation
Downloads
Results for foxa3
Z-value: 0.75
Transcription factors associated with foxa3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxa3
|
ENSDARG00000012788 | forkhead box A3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxa3 | dr10_dc_chr18_-_46356213_46356267 | 0.67 | 4.8e-03 | Click! |
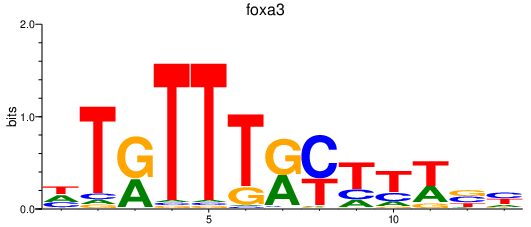
Activity profile of foxa3 motif
Sorted Z-values of foxa3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of foxa3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_51392394 | 2.67 |
ENSDART00000143276
|
setbp1
|
SET binding protein 1 |
| chr11_+_7518953 | 2.43 |
ENSDART00000171813
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr16_+_24063849 | 2.29 |
ENSDART00000135084
|
apoa2
|
apolipoprotein A-II |
| chr25_-_34677352 | 2.04 |
ENSDART00000115210
|
ano9a
|
anoctamin 9a |
| chr9_+_54185607 | 1.88 |
ENSDART00000165991
|
lect1
|
leukocyte cell derived chemotaxin 1 |
| chr5_+_15880920 | 1.70 |
ENSDART00000168643
|
CABZ01088700.1
|
ENSDARG00000101452 |
| chr11_-_29316162 | 1.70 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr15_+_37039861 | 1.63 |
ENSDART00000172664
|
kirrel3l
|
kin of IRRE like 3 like |
| chr16_+_23999029 | 1.61 |
ENSDART00000129525
|
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr7_-_32752138 | 1.54 |
ENSDART00000052383
|
cd81a
|
CD81 molecule a |
| chr9_-_45802484 | 1.53 |
ENSDART00000139019
|
agr1
|
anterior gradient 1 |
| chr4_-_73485204 | 1.51 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| chr2_+_5651428 | 1.41 |
ENSDART00000111220
|
mb21d2
|
Mab-21 domain containing 2 |
| chr5_+_45822196 | 1.39 |
|
|
|
| chr12_+_36796886 | 1.33 |
ENSDART00000125900
|
hs3st3b1b
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1b |
| chr1_-_10423289 | 1.30 |
|
|
|
| chr25_-_30845998 | 1.23 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr4_-_13922285 | 1.22 |
ENSDART00000080334
|
yaf2
|
YY1 associated factor 2 |
| chr2_+_30932612 | 1.18 |
ENSDART00000132450
ENSDART00000137012 |
myom1a
|
myomesin 1a (skelemin) |
| chr4_-_9721600 | 1.16 |
ENSDART00000067190
|
tspan9b
|
tetraspanin 9b |
| chr19_-_29205158 | 1.15 |
ENSDART00000026992
|
sox4a
|
SRY (sex determining region Y)-box 4a |
| chr23_-_20332076 | 1.13 |
ENSDART00000147326
|
lamb2
|
laminin, beta 2 (laminin S) |
| chr5_+_31745031 | 1.09 |
ENSDART00000147132
|
c9
|
complement component 9 |
| chr21_-_39035904 | 1.08 |
ENSDART00000075935
|
vtnb
|
vitronectin b |
| chr16_-_30606364 | 1.08 |
ENSDART00000147986
|
lmna
|
lamin A |
| chr5_+_49093250 | 1.07 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| KN150702v1_+_177145 | 1.07 |
ENSDART00000166225
|
CABZ01076674.1
|
ENSDARG00000102620 |
| chr7_-_57796486 | 1.06 |
ENSDART00000043984
|
ank2b
|
ankyrin 2b, neuronal |
| chr21_+_17731439 | 0.99 |
ENSDART00000124173
|
rxraa
|
retinoid X receptor, alpha a |
| chr24_+_9335428 | 0.97 |
ENSDART00000132688
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr16_-_8749368 | 0.96 |
ENSDART00000162988
|
cobl
|
cordon-bleu WH2 repeat protein |
| chr4_+_1750689 | 0.95 |
ENSDART00000146779
|
slc38a2
|
solute carrier family 38, member 2 |
| chr8_+_49582361 | 0.95 |
ENSDART00000108613
|
rasef
|
RAS and EF-hand domain containing |
| chr20_-_42306410 | 0.94 |
ENSDART00000074959
|
slc35f1
|
solute carrier family 35, member F1 |
| chr5_+_49093134 | 0.92 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr15_-_25430003 | 0.91 |
ENSDART00000152651
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr8_-_16662185 | 0.91 |
ENSDART00000076542
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr14_+_38375351 | 0.90 |
ENSDART00000175744
|
BX088707.3
|
ENSDARG00000107351 |
| chr11_-_30387031 | 0.89 |
|
|
|
| chr21_-_1600125 | 0.86 |
ENSDART00000066621
|
vmo1b
|
vitelline membrane outer layer 1 homolog b |
| chr5_+_35913418 | 0.85 |
ENSDART00000013590
|
wnt11
|
wingless-type MMTV integration site family, member 11 |
| chr20_-_9992898 | 0.85 |
ENSDART00000080664
|
ACTC1
|
zgc:86709 |
| chr10_+_7043530 | 0.85 |
|
|
|
| chr10_+_35535209 | 0.85 |
ENSDART00000109705
|
phldb2a
|
pleckstrin homology-like domain, family B, member 2a |
| chr1_+_50395721 | 0.84 |
ENSDART00000134065
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr21_-_5852663 | 0.84 |
ENSDART00000130521
|
CACFD1
|
calcium channel flower domain containing 1 |
| chr8_+_39762186 | 0.83 |
ENSDART00000143413
|
si:ch211-170d8.2
|
si:ch211-170d8.2 |
| chr2_+_38178934 | 0.82 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr4_+_10722966 | 0.82 |
ENSDART00000102534
ENSDART00000067253 ENSDART00000136000 |
stab2
|
stabilin 2 |
| chr8_+_17148864 | 0.81 |
ENSDART00000140531
|
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr9_-_6683651 | 0.80 |
ENSDART00000061593
|
pou3f3a
|
POU class 3 homeobox 3a |
| chr9_-_22394674 | 0.75 |
ENSDART00000101869
|
crygm2d12
|
crystallin, gamma M2d12 |
| chr23_-_44729542 | 0.74 |
ENSDART00000024082
|
psmb6
|
proteasome subunit beta 6 |
| chr1_+_25378105 | 0.73 |
ENSDART00000059264
|
mxd4
|
MAX dimerization protein 4 |
| chr5_+_41492032 | 0.72 |
ENSDART00000171678
|
ubb
|
ubiquitin B |
| chr5_-_43121949 | 0.72 |
ENSDART00000157093
|
si:dkey-40c11.1
|
si:dkey-40c11.1 |
| chr9_+_52872108 | 0.71 |
ENSDART00000157453
ENSDART00000163684 |
nme8
|
NME/NM23 family member 8 |
| chr10_+_32007448 | 0.71 |
ENSDART00000019416
|
lhfp
|
lipoma HMGIC fusion partner |
| chr16_+_5466342 | 0.71 |
ENSDART00000160008
|
plecb
|
plectin b |
| chr24_+_20430778 | 0.71 |
ENSDART00000010488
|
klhl40b
|
kelch-like family member 40b |
| chr22_+_31866783 | 0.69 |
ENSDART00000159825
|
dock3
|
dedicator of cytokinesis 3 |
| chr7_-_26572828 | 0.69 |
ENSDART00000162241
|
si:ch211-107p11.3
|
si:ch211-107p11.3 |
| chr14_-_6624878 | 0.68 |
ENSDART00000166439
|
si:ch211-266k2.1
|
si:ch211-266k2.1 |
| chr3_+_5665300 | 0.68 |
ENSDART00000101807
|
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr1_+_1657493 | 0.67 |
ENSDART00000103850
|
atp1a1a.3
|
ATPase, Na+/K+ transporting, alpha 1a polypeptide, tandem duplicate 3 |
| chr15_+_7179382 | 0.66 |
ENSDART00000101578
|
her8.2
|
hairy-related 8.2 |
| chr16_+_21098948 | 0.65 |
ENSDART00000006043
|
hoxa11b
|
homeobox A11b |
| chr16_-_21981065 | 0.65 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr22_-_36915567 | 0.64 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr3_-_57691670 | 0.64 |
ENSDART00000156522
|
cant1a
|
calcium activated nucleotidase 1a |
| chr6_-_56013025 | 0.64 |
ENSDART00000011652
|
eya2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr6_+_10491884 | 0.64 |
ENSDART00000151017
|
BX936397.2
|
ENSDARG00000096373 |
| chr15_-_44029439 | 0.64 |
|
|
|
| chr1_+_25974174 | 0.62 |
ENSDART00000152144
|
si:dkey-25o16.2
|
si:dkey-25o16.2 |
| chr16_-_54634679 | 0.62 |
ENSDART00000075275
ENSDART00000115024 |
pklr
|
pyruvate kinase, liver and RBC |
| chr4_+_13453879 | 0.61 |
ENSDART00000145069
|
dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr4_-_14927871 | 0.61 |
ENSDART00000110199
|
prdm4
|
PR domain containing 4 |
| chr11_-_21422588 | 0.61 |
|
|
|
| chr4_-_16461748 | 0.60 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| chr18_-_48375325 | 0.60 |
|
|
|
| chr7_+_13750329 | 0.60 |
ENSDART00000091470
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr5_+_34022151 | 0.60 |
ENSDART00000141338
|
enc1
|
ectodermal-neural cortex 1 |
| chr13_-_11535112 | 0.59 |
ENSDART00000102411
|
dctn1b
|
dynactin 1b |
| chr6_+_54231519 | 0.58 |
ENSDART00000149542
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr16_-_41181493 | 0.58 |
ENSDART00000102649
ENSDART00000145956 |
ptpn23a
|
protein tyrosine phosphatase, non-receptor type 23, a |
| chr24_+_9335316 | 0.58 |
ENSDART00000132688
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr15_-_16271208 | 0.58 |
|
|
|
| chr9_+_54185729 | 0.57 |
ENSDART00000165991
|
lect1
|
leukocyte cell derived chemotaxin 1 |
| chr4_+_119964 | 0.57 |
ENSDART00000177177
ENSDART00000159996 |
tbk1
|
TANK-binding kinase 1 |
| chr5_+_26312951 | 0.55 |
ENSDART00000126609
|
tbx3b
|
T-box 3b |
| chr6_-_40660116 | 0.55 |
ENSDART00000154359
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr3_+_32360862 | 0.54 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr4_+_14361655 | 0.54 |
ENSDART00000007103
|
nuak1a
|
NUAK family, SNF1-like kinase, 1a |
| chr21_-_14534576 | 0.54 |
ENSDART00000089967
ENSDART00000132142 |
cacna1bb
|
calcium channel, voltage-dependent, N type, alpha 1B subunit, b |
| chr21_-_25258473 | 0.54 |
ENSDART00000101153
ENSDART00000147860 ENSDART00000087910 |
st14b
|
suppression of tumorigenicity 14 (colon carcinoma) b |
| chr16_-_31670211 | 0.54 |
ENSDART00000138216
|
CR855311.1
|
ENSDARG00000090352 |
| chr19_+_1922930 | 0.54 |
ENSDART00000013217
|
snrpd1
|
small nuclear ribonucleoprotein D1 polypeptide |
| chr6_+_24717985 | 0.53 |
ENSDART00000165609
|
barhl2
|
BarH-like homeobox 2 |
| chr7_+_38123920 | 0.53 |
ENSDART00000138669
|
cep89
|
centrosomal protein 89 |
| chr7_-_58427369 | 0.52 |
ENSDART00000149347
|
chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr18_+_14369543 | 0.51 |
|
|
|
| chr18_+_17622557 | 0.50 |
ENSDART00000046891
|
cetp
|
cholesteryl ester transfer protein, plasma |
| chr24_-_16994956 | 0.50 |
ENSDART00000111079
|
CR376737.1
|
ENSDARG00000077318 |
| chr2_+_20814888 | 0.49 |
ENSDART00000170793
|
agla
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase a |
| chr1_+_44239727 | 0.48 |
ENSDART00000141145
|
si:dkey-9i23.16
|
si:dkey-9i23.16 |
| chr23_-_30772173 | 0.48 |
ENSDART00000138841
|
CR456642.1
|
ENSDARG00000092307 |
| chr14_+_7626822 | 0.48 |
ENSDART00000109941
|
cxxc5b
|
CXXC finger protein 5b |
| chr8_-_3253562 | 0.47 |
ENSDART00000035965
|
fut9b
|
fucosyltransferase 9b |
| chr15_+_36329506 | 0.47 |
ENSDART00000099501
|
masp1
|
mannan-binding lectin serine peptidase 1 |
| chr16_-_30816954 | 0.46 |
ENSDART00000129594
|
ptk2ab
|
protein tyrosine kinase 2ab |
| chr6_-_31160132 | 0.46 |
ENSDART00000153592
|
CR847887.1
|
ENSDARG00000097226 |
| chr4_-_885413 | 0.46 |
ENSDART00000022668
|
aim1b
|
crystallin beta-gamma domain containing 1b |
| chr11_-_36088889 | 0.46 |
ENSDART00000146495
|
psma5
|
proteasome subunit alpha 5 |
| chr16_+_11833310 | 0.46 |
ENSDART00000060266
|
ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr22_-_14103803 | 0.45 |
ENSDART00000062902
|
ENSDARG00000042857
|
ENSDARG00000042857 |
| chr13_+_5850284 | 0.45 |
ENSDART00000121598
|
phf10
|
PHD finger protein 10 |
| chr3_+_25888520 | 0.45 |
ENSDART00000135389
|
foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr5_-_25509188 | 0.45 |
ENSDART00000137376
|
BX510923.1
|
ENSDARG00000094488 |
| chr5_-_25509259 | 0.44 |
ENSDART00000137376
|
BX510923.1
|
ENSDARG00000094488 |
| chr17_+_24428093 | 0.44 |
ENSDART00000131417
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr1_-_44239584 | 0.44 |
ENSDART00000137216
|
tmem176
|
transmembrane protein 176 |
| chr3_-_15475774 | 0.43 |
ENSDART00000136890
|
BX682550.1
|
ENSDARG00000090026 |
| chr1_+_25974337 | 0.43 |
ENSDART00000152617
|
si:dkey-25o16.2
|
si:dkey-25o16.2 |
| chr12_+_26376471 | 0.43 |
ENSDART00000022495
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr2_-_56896934 | 0.43 |
ENSDART00000164086
|
slc25a42
|
solute carrier family 25, member 42 |
| chr8_-_13182911 | 0.42 |
ENSDART00000090541
|
si:ch73-61d6.3
|
si:ch73-61d6.3 |
| chr1_-_25377787 | 0.41 |
|
|
|
| chr18_-_16806665 | 0.41 |
ENSDART00000048722
|
ampd3b
|
adenosine monophosphate deaminase 3b |
| chr21_+_30465250 | 0.41 |
ENSDART00000043727
|
slc7a3a
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3a |
| chr17_+_32547862 | 0.41 |
ENSDART00000018423
|
ywhaqb
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide b |
| chr12_+_19240407 | 0.41 |
ENSDART00000041711
|
gspt1
|
G1 to S phase transition 1 |
| chr22_-_14103942 | 0.41 |
ENSDART00000140323
|
ENSDARG00000042857
|
ENSDARG00000042857 |
| chr6_+_12619062 | 0.41 |
ENSDART00000156341
|
fam117ba
|
family with sequence similarity 117, member Ba |
| chr15_+_29190162 | 0.41 |
ENSDART00000105222
|
zgc:101731
|
zgc:101731 |
| chr2_-_6351592 | 0.40 |
|
|
|
| chr13_-_40600924 | 0.40 |
ENSDART00000099847
ENSDART00000057046 |
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr10_-_14587464 | 0.40 |
ENSDART00000057865
|
ier3ip1
|
immediate early response 3 interacting protein 1 |
| chr9_+_51733862 | 0.40 |
ENSDART00000137426
|
gcgb
|
glucagon b |
| chr8_-_27839244 | 0.40 |
ENSDART00000136562
|
cttnbp2nlb
|
CTTNBP2 N-terminal like b |
| chr1_-_22144014 | 0.39 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr15_-_31236277 | 0.39 |
|
|
|
| chr1_+_44239682 | 0.39 |
ENSDART00000141145
|
si:dkey-9i23.16
|
si:dkey-9i23.16 |
| chr4_-_6558687 | 0.39 |
ENSDART00000150774
|
foxp2
|
forkhead box P2 |
| chr1_+_25974264 | 0.39 |
ENSDART00000152785
|
si:dkey-25o16.2
|
si:dkey-25o16.2 |
| chr8_+_50498398 | 0.38 |
|
|
|
| chr24_+_9124068 | 0.38 |
|
|
|
| chr14_+_27811744 | 0.38 |
|
|
|
| KN150520v1_+_33937 | 0.38 |
|
|
|
| chr14_-_29459799 | 0.38 |
ENSDART00000005568
|
pdlim3b
|
PDZ and LIM domain 3b |
| chr2_-_57133471 | 0.38 |
|
|
|
| chr11_+_7518748 | 0.37 |
ENSDART00000171813
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr5_-_25509310 | 0.37 |
ENSDART00000137376
|
BX510923.1
|
ENSDARG00000094488 |
| chr20_+_15119754 | 0.37 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr21_+_30512356 | 0.37 |
ENSDART00000109719
|
rab38c
|
RAB38c, member of RAS oncogene family |
| chr20_+_38768950 | 0.37 |
ENSDART00000146544
|
mpv17
|
MpV17 mitochondrial inner membrane protein |
| chr20_+_15119519 | 0.36 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr20_+_52740555 | 0.36 |
ENSDART00000110777
|
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr10_+_9762039 | 0.36 |
|
|
|
| chr12_-_304698 | 0.35 |
|
|
|
| chr8_-_7547623 | 0.35 |
ENSDART00000178825
|
hcfc1b
|
host cell factor C1b |
| chr5_-_43122200 | 0.35 |
ENSDART00000157093
|
si:dkey-40c11.1
|
si:dkey-40c11.1 |
| chr5_+_18826505 | 0.35 |
|
|
|
| chr15_+_37040064 | 0.35 |
ENSDART00000172664
|
kirrel3l
|
kin of IRRE like 3 like |
| chr5_+_18826621 | 0.35 |
|
|
|
| chr5_-_32282404 | 0.34 |
ENSDART00000166698
|
lrsam1
|
leucine rich repeat and sterile alpha motif containing 1 |
| chr23_+_42920817 | 0.34 |
ENSDART00000055577
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr2_-_16634647 | 0.34 |
|
|
|
| chr4_-_18447913 | 0.34 |
ENSDART00000141671
|
socs2
|
suppressor of cytokine signaling 2 |
| chr8_+_7974156 | 0.34 |
|
|
|
| chr9_+_28113358 | 0.33 |
|
|
|
| chr8_-_14146621 | 0.33 |
ENSDART00000063817
|
ndufb11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11 |
| chr4_+_8204477 | 0.32 |
ENSDART00000176361
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr20_+_29307142 | 0.32 |
ENSDART00000153016
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr13_+_42417935 | 0.32 |
ENSDART00000145409
|
si:dkey-221j11.3
|
si:dkey-221j11.3 |
| chr14_+_20862466 | 0.32 |
ENSDART00000059796
|
ENSDARG00000019213
|
ENSDARG00000019213 |
| chr15_+_21399127 | 0.32 |
ENSDART00000154914
|
BX663520.1
|
ENSDARG00000097950 |
| chr15_+_21399299 | 0.31 |
ENSDART00000154914
|
BX663520.1
|
ENSDARG00000097950 |
| chr23_+_29431173 | 0.31 |
ENSDART00000027255
|
tardbpl
|
TAR DNA binding protein, like |
| chr9_+_6684835 | 0.31 |
|
|
|
| chr17_+_24018420 | 0.31 |
ENSDART00000140767
|
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr23_-_38228305 | 0.30 |
ENSDART00000131791
|
pfdn4
|
prefoldin subunit 4 |
| chr10_+_10770717 | 0.30 |
ENSDART00000146910
|
swi5
|
SWI5 homologous recombination repair protein |
| chr4_-_73485366 | 0.30 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| chr20_+_29307039 | 0.30 |
ENSDART00000152949
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr17_-_7990960 | 0.29 |
|
|
|
| chr6_-_47928632 | 0.29 |
|
|
|
| chr4_+_13453584 | 0.28 |
ENSDART00000145069
|
dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr9_-_33330725 | 0.28 |
|
|
|
| chr16_-_12095302 | 0.28 |
ENSDART00000148666
ENSDART00000029121 |
usp5
|
ubiquitin specific peptidase 5 (isopeptidase T) |
| chr15_+_6870469 | 0.28 |
|
|
|
| chr5_-_22847399 | 0.28 |
ENSDART00000169258
|
nlgn3b
|
neuroligin 3b |
| chr5_-_11444117 | 0.27 |
ENSDART00000165446
|
rnft2
|
ring finger protein, transmembrane 2 |
| chr21_-_26453406 | 0.27 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr4_-_73485124 | 0.27 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| chr22_-_36915720 | 0.27 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr2_+_30911052 | 0.26 |
ENSDART00000132645
|
chmp5a
|
charged multivesicular body protein 5a |
| chr25_+_33924376 | 0.26 |
|
|
|
| chr16_-_8367090 | 0.25 |
ENSDART00000129068
|
entpd3
|
ectonucleoside triphosphate diphosphohydrolase 3 |
| chr2_-_31817355 | 0.25 |
ENSDART00000112763
|
retreg1
|
reticulophagy regulator 1 |
| chr6_-_43782889 | 0.23 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.3 | 1.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.2 | 0.7 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.2 | 1.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 1.5 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.2 | 1.1 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.2 | 0.8 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.2 | 0.9 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 0.5 | GO:0002706 | regulation of lymphocyte mediated immunity(GO:0002706) |
| 0.2 | 2.0 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 0.2 | 0.8 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.2 | 0.5 | GO:0034372 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) reverse cholesterol transport(GO:0043691) phospholipid homeostasis(GO:0055091) |
| 0.2 | 1.1 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.2 | 1.7 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.2 | 1.2 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.1 | 2.0 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.1 | 1.0 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 1.0 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 0.4 | GO:0015822 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.1 | 0.4 | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.1 | 0.4 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.6 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.8 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 1.1 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.3 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.3 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.1 | 1.2 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 0.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.2 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.1 | 0.7 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.1 | 1.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.5 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.2 | GO:0046874 | quinolinate biosynthetic process(GO:0019805) quinolinate metabolic process(GO:0046874) |
| 0.0 | 2.8 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.9 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.0 | 0.5 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.7 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.4 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.2 | GO:0048937 | lateral line nerve glial cell differentiation(GO:0048895) myelination of lateral line nerve axons(GO:0048897) posterior lateral line nerve glial cell differentiation(GO:0048931) myelination of posterior lateral line nerve axons(GO:0048932) lateral line nerve glial cell development(GO:0048937) lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048938) posterior lateral line nerve glial cell development(GO:0048941) posterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048942) |
| 0.0 | 1.1 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.3 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 1.0 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:0006699 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.0 | 0.4 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.2 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.3 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.6 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 1.7 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 0.4 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.9 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.3 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.0 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 1.5 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.1 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.7 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.5 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.0 | 0.4 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 1.2 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.6 | GO:0003205 | cardiac chamber development(GO:0003205) |
| 0.0 | 0.8 | GO:0002088 | lens development in camera-type eye(GO:0002088) visual perception(GO:0007601) |
| 0.0 | 0.6 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.1 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.1 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 1.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.5 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.1 | 1.2 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.1 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 0.3 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.1 | 0.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 1.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.5 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 0.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.1 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 0.2 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 1.3 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 2.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 2.8 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 1.1 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.4 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.5 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 1.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 0.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 1.0 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 1.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.4 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 0.4 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 0.4 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.1 | 1.0 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.9 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.5 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 1.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.1 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.3 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.1 | 0.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.7 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.0 | 0.4 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.9 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.7 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0048270 | methionine adenosyltransferase regulator activity(GO:0048270) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 3.9 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 2.0 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.5 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.5 | GO:0099604 | ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.3 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 2.0 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.4 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 1.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 2.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 1.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 0.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 0.6 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 0.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.2 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.3 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.5 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.6 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |