Project
DANIO-CODE
Navigation
Downloads
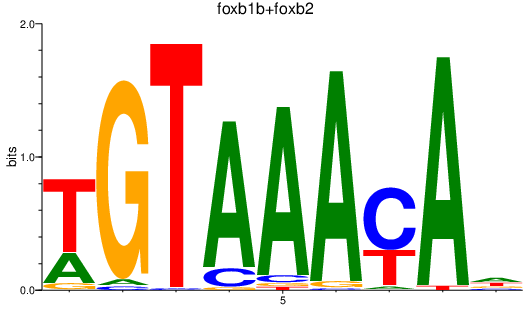
Results for foxb1b+foxb2
Z-value: 1.44
Transcription factors associated with foxb1b+foxb2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxb2
|
ENSDARG00000037475 | forkhead box B2 |
|
foxb1b
|
ENSDARG00000053650 | forkhead box B1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxb1b | dr10_dc_chr7_+_29189933_29189974 | -0.58 | 1.9e-02 | Click! |
Activity profile of foxb1b+foxb2 motif
Sorted Z-values of foxb1b+foxb2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of foxb1b+foxb2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_32636372 | 6.16 |
ENSDART00000085512
ENSDART00000144694 |
kank1b
|
KN motif and ankyrin repeat domains 1b |
| chr8_+_37716781 | 5.20 |
ENSDART00000108556
|
npm2a
|
nucleophosmin/nucleoplasmin, 2a |
| chr4_-_14208573 | 4.34 |
ENSDART00000015134
|
twf1b
|
twinfilin actin-binding protein 1b |
| chr3_+_42383724 | 3.24 |
|
|
|
| chr11_-_25019899 | 3.09 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr24_-_14446593 | 2.93 |
|
|
|
| chr7_-_37283707 | 2.83 |
ENSDART00000148905
ENSDART00000150229 |
cylda
|
cylindromatosis (turban tumor syndrome), a |
| chr11_-_6442588 | 2.79 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr14_-_7102535 | 2.75 |
ENSDART00000036463
|
dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr24_-_20999883 | 2.59 |
ENSDART00000155652
|
gramd1c
|
GRAM domain containing 1c |
| chr7_+_45747622 | 2.45 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr7_+_45747395 | 2.30 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr5_-_21523599 | 2.28 |
ENSDART00000023306
|
asb12a
|
ankyrin repeat and SOCS box-containing 12a |
| chr16_-_47446494 | 2.27 |
ENSDART00000032188
|
si:dkey-256h2.1
|
si:dkey-256h2.1 |
| chr5_-_68011971 | 2.20 |
ENSDART00000141699
|
mepce
|
methylphosphate capping enzyme |
| chr16_-_25318413 | 2.11 |
ENSDART00000058943
|
ENSDARG00000040280
|
ENSDARG00000040280 |
| chr3_-_20944579 | 2.09 |
ENSDART00000153739
|
nlk1
|
nemo-like kinase, type 1 |
| chr3_+_25776714 | 2.02 |
ENSDART00000170324
|
tom1
|
target of myb1 membrane trafficking protein |
| chr14_+_14850200 | 2.01 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr8_+_26015379 | 1.96 |
ENSDART00000142555
|
arih2
|
ariadne homolog 2 (Drosophila) |
| chr6_-_31377513 | 1.92 |
ENSDART00000145715
|
ak4
|
adenylate kinase 4 |
| chr9_-_21677941 | 1.91 |
ENSDART00000121939
ENSDART00000080404 |
mphosph8
|
M-phase phosphoprotein 8 |
| chr23_-_16755868 | 1.90 |
ENSDART00000020810
|
sdcbp2
|
syndecan binding protein (syntenin) 2 |
| chr5_+_36049836 | 1.89 |
ENSDART00000147667
|
alkbh6
|
alkB homolog 6 |
| chr1_+_21244200 | 1.87 |
ENSDART00000087729
|
kdm4c
|
lysine (K)-specific demethylase 4C |
| chr25_-_7794186 | 1.83 |
ENSDART00000104686
|
ambra1b
|
autophagy/beclin-1 regulator 1b |
| chr17_+_43916865 | 1.82 |
ENSDART00000049503
|
peli2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr19_+_32670271 | 1.82 |
|
|
|
| chr2_-_54224744 | 1.81 |
|
|
|
| chr24_+_26223767 | 1.81 |
|
|
|
| chr6_+_11162062 | 1.79 |
ENSDART00000151548
|
senp2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr15_-_33342455 | 1.78 |
|
|
|
| chr16_-_5205600 | 1.77 |
ENSDART00000148955
|
bckdhb
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr2_-_29939721 | 1.76 |
ENSDART00000031130
|
paxip1
|
PAX interacting (with transcription-activation domain) protein 1 |
| chr12_-_19063761 | 1.75 |
ENSDART00000153343
|
zc3h7b
|
zinc finger CCCH-type containing 7B |
| chr23_-_1562572 | 1.74 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr5_-_17372745 | 1.69 |
ENSDART00000141978
|
si:dkey-112e17.1
|
si:dkey-112e17.1 |
| chr11_-_14044356 | 1.68 |
ENSDART00000085158
|
tmem259
|
transmembrane protein 259 |
| chr23_+_20586745 | 1.67 |
ENSDART00000157522
|
dpm1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr17_-_9806363 | 1.66 |
ENSDART00000021942
|
eapp
|
e2f-associated phosphoprotein |
| chr5_+_61136434 | 1.66 |
ENSDART00000113508
|
hnrnpul1l
|
heterogeneous nuclear ribonucleoprotein U-like 1 like |
| chr17_+_31205274 | 1.64 |
ENSDART00000155580
|
ccdc32
|
coiled-coil domain containing 32 |
| chr8_+_7655257 | 1.63 |
ENSDART00000170184
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr19_+_32000438 | 1.62 |
ENSDART00000150910
|
gmnn
|
geminin, DNA replication inhibitor |
| chr8_-_3974320 | 1.60 |
ENSDART00000159895
|
mtmr3
|
myotubularin related protein 3 |
| chr18_+_14725283 | 1.59 |
ENSDART00000146128
|
uri1
|
URI1, prefoldin-like chaperone |
| chr7_-_8466131 | 1.52 |
ENSDART00000065488
|
tex261
|
testis expressed 261 |
| chr9_-_3548174 | 1.51 |
ENSDART00000145043
ENSDART00000169586 |
dcaf17
|
ddb1 and cul4 associated factor 17 |
| chr13_-_25711537 | 1.51 |
ENSDART00000015154
|
papolg
|
poly(A) polymerase gamma |
| chr25_+_30627892 | 1.50 |
|
|
|
| chr25_+_30627755 | 1.50 |
|
|
|
| chr18_-_5167807 | 1.50 |
|
|
|
| chr11_-_25019298 | 1.48 |
ENSDART00000130477
ENSDART00000079578 |
snai1a
|
snail family zinc finger 1a |
| chr23_+_44470297 | 1.46 |
ENSDART00000149842
|
MEPCE
|
methylphosphate capping enzyme |
| chr7_+_15197911 | 1.46 |
ENSDART00000046542
|
igf1rb
|
insulin-like growth factor 1b receptor |
| chr23_+_22052173 | 1.45 |
ENSDART00000087110
|
eif4g3b
|
eukaryotic translation initiation factor 4 gamma, 3b |
| chr14_+_20941526 | 1.43 |
ENSDART00000138551
|
smim19
|
small integral membrane protein 19 |
| chr16_-_25317921 | 1.42 |
|
|
|
| chr15_-_5913264 | 1.42 |
ENSDART00000155156
ENSDART00000155971 |
si:ch73-281n10.2
|
si:ch73-281n10.2 |
| chr20_-_53123124 | 1.42 |
ENSDART00000162812
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr3_-_20944854 | 1.41 |
ENSDART00000109790
|
nlk1
|
nemo-like kinase, type 1 |
| chr12_-_7201358 | 1.41 |
ENSDART00000048866
|
ipmkb
|
inositol polyphosphate multikinase b |
| chr2_-_48103519 | 1.41 |
ENSDART00000056305
|
fzd8b
|
frizzled class receptor 8b |
| chr25_-_28630138 | 1.40 |
|
|
|
| chr22_-_21873054 | 1.37 |
|
|
|
| chr24_+_12693997 | 1.37 |
|
|
|
| chr5_-_47610384 | 1.35 |
|
|
|
| chr14_-_6901209 | 1.35 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr16_-_52090636 | 1.34 |
ENSDART00000155308
|
FO704712.1
|
ENSDARG00000063300 |
| chr22_-_4787016 | 1.34 |
ENSDART00000140313
|
tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr20_-_25587274 | 1.33 |
ENSDART00000141340
|
si:dkey-183n20.15
|
si:dkey-183n20.15 |
| chr19_-_19452999 | 1.33 |
ENSDART00000163359
ENSDART00000167951 |
dync1li1
|
dynein, cytoplasmic 1, light intermediate chain 1 |
| chr17_-_30635487 | 1.32 |
ENSDART00000154960
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr3_-_26060787 | 1.31 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr20_+_30476065 | 1.31 |
ENSDART00000062441
|
rnaseh1
|
ribonuclease H1 |
| chr1_+_53294559 | 1.29 |
ENSDART00000159900
|
ccsapa
|
centriole, cilia and spindle-associated protein a |
| chr23_+_22052045 | 1.29 |
ENSDART00000145172
|
eif4g3b
|
eukaryotic translation initiation factor 4 gamma, 3b |
| chr21_-_25991445 | 1.28 |
ENSDART00000034875
|
sdf2
|
stromal cell-derived factor 2 |
| chr17_+_32408151 | 1.28 |
ENSDART00000155519
|
si:ch211-139d20.3
|
si:ch211-139d20.3 |
| chr21_+_20350218 | 1.28 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr25_-_3091345 | 1.28 |
ENSDART00000032390
|
gtf2h1
|
general transcription factor IIH, polypeptide 1 |
| chr25_-_10694465 | 1.27 |
ENSDART00000127054
|
BX572619.1
|
ENSDARG00000089303 |
| chr22_-_35003371 | 1.26 |
ENSDART00000133537
|
arhgap19
|
Rho GTPase activating protein 19 |
| chr8_+_7655310 | 1.26 |
ENSDART00000170184
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr7_-_31346802 | 1.24 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr7_+_66660882 | 1.24 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr23_-_3815871 | 1.24 |
ENSDART00000137826
|
hmga1a
|
high mobility group AT-hook 1a |
| chr17_+_10592507 | 1.24 |
ENSDART00000097274
|
ATG14
|
autophagy related 14 |
| chr15_+_20054306 | 1.24 |
ENSDART00000155199
|
zgc:112083
|
zgc:112083 |
| chr13_+_40689102 | 1.23 |
ENSDART00000016960
|
prkg1a
|
protein kinase, cGMP-dependent, type Ia |
| chr24_+_19065679 | 1.23 |
ENSDART00000139299
|
zgc:162928
|
zgc:162928 |
| chr4_-_12782246 | 1.23 |
ENSDART00000058020
|
helb
|
helicase (DNA) B |
| chr24_+_14095601 | 1.23 |
ENSDART00000124740
|
ncoa2
|
nuclear receptor coactivator 2 |
| chr4_+_8669695 | 1.23 |
ENSDART00000168768
|
adipor2
|
adiponectin receptor 2 |
| chr10_-_39362315 | 1.22 |
ENSDART00000023831
|
cry5
|
cryptochrome circadian clock 5 |
| chr19_+_5399535 | 1.22 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr21_-_25991335 | 1.21 |
ENSDART00000034875
|
sdf2
|
stromal cell-derived factor 2 |
| chr11_+_11136919 | 1.21 |
ENSDART00000026135
|
ly75
|
lymphocyte antigen 75 |
| chr11_-_30261689 | 1.20 |
ENSDART00000101667
|
map4k3a
|
mitogen-activated protein kinase kinase kinase kinase 3a |
| chr5_-_66070385 | 1.20 |
ENSDART00000032909
|
kat5b
|
K(lysine) acetyltransferase 5b |
| chr13_+_36554762 | 1.19 |
ENSDART00000136030
|
atp5s
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit s |
| chr25_-_7794262 | 1.19 |
ENSDART00000156761
|
ambra1b
|
autophagy/beclin-1 regulator 1b |
| chr3_+_13780104 | 1.18 |
ENSDART00000164179
|
syce2
|
synaptonemal complex central element protein 2 |
| chr14_-_21816253 | 1.18 |
ENSDART00000113752
|
si:dkey-6i22.5
|
si:dkey-6i22.5 |
| chr5_-_64432697 | 1.18 |
ENSDART00000165556
|
tor2a
|
torsin family 2, member A |
| chr11_-_25495823 | 1.17 |
ENSDART00000171935
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr21_-_26369503 | 1.16 |
ENSDART00000137312
|
eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr6_-_43297126 | 1.16 |
ENSDART00000154170
|
frmd4ba
|
FERM domain containing 4Ba |
| chr12_+_35855929 | 1.16 |
|
|
|
| chr3_-_29837411 | 1.15 |
ENSDART00000139310
|
bcat2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr12_+_47828020 | 1.15 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr25_-_25339397 | 1.14 |
ENSDART00000171589
|
hrasa
|
v-Ha-ras Harvey rat sarcoma viral oncogene homolog a |
| chr8_+_10825036 | 1.14 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr16_-_26947117 | 1.14 |
ENSDART00000111156
|
pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr21_+_3764216 | 1.14 |
ENSDART00000149777
|
setx
|
senataxin |
| chr5_-_65564733 | 1.14 |
ENSDART00000178162
|
CABZ01049025.1
|
ENSDARG00000107190 |
| chr12_+_30611399 | 1.14 |
ENSDART00000153036
|
slc35f3a
|
solute carrier family 35, member F3a |
| chr21_-_25991595 | 1.14 |
ENSDART00000034875
|
sdf2
|
stromal cell-derived factor 2 |
| chr3_+_56882443 | 1.13 |
|
|
|
| chr2_-_57826189 | 1.13 |
ENSDART00000131420
|
si:dkeyp-68b7.5
|
si:dkeyp-68b7.5 |
| chr16_+_46443800 | 1.12 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr9_+_28329384 | 1.12 |
ENSDART00000046880
|
plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| chr9_-_3548211 | 1.10 |
ENSDART00000145043
ENSDART00000169586 |
dcaf17
|
ddb1 and cul4 associated factor 17 |
| chr7_-_8465929 | 1.09 |
ENSDART00000172928
|
tex261
|
testis expressed 261 |
| chr16_-_25318334 | 1.09 |
ENSDART00000058943
|
ENSDARG00000040280
|
ENSDARG00000040280 |
| chr3_+_52745606 | 1.08 |
ENSDART00000104683
|
pbx4
|
pre-B-cell leukemia transcription factor 4 |
| chr16_+_41004372 | 1.08 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr8_-_25696708 | 1.08 |
ENSDART00000007482
|
tspy
|
testis specific protein, Y-linked |
| chr6_-_52725402 | 1.07 |
ENSDART00000033949
|
oser1
|
oxidative stress responsive serine-rich 1 |
| chr5_+_51979524 | 1.07 |
ENSDART00000168317
|
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr7_+_59371957 | 1.07 |
ENSDART00000125570
|
trmt44
|
tRNA methyltransferase 44 homolog (S. cerevisiae) |
| chr16_+_25381315 | 1.07 |
ENSDART00000114528
|
tbc1d31
|
TBC1 domain family, member 31 |
| chr18_+_5066229 | 1.07 |
ENSDART00000157671
|
CABZ01080599.1
|
ENSDARG00000100626 |
| chr16_-_39317068 | 1.06 |
ENSDART00000133642
|
gpd1l
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr8_+_7815256 | 1.06 |
ENSDART00000165575
|
cxxc1a
|
CXXC finger protein 1a |
| chr4_-_14625618 | 1.06 |
ENSDART00000137847
|
plxnb2a
|
plexin b2a |
| chr11_+_18450008 | 1.06 |
ENSDART00000162694
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr20_-_53123015 | 1.05 |
ENSDART00000162812
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr23_-_36349563 | 1.05 |
|
|
|
| chr15_-_28547372 | 1.05 |
ENSDART00000057696
|
git1
|
G protein-coupled receptor kinase interacting ArfGAP 1 |
| chr13_-_36554590 | 1.04 |
ENSDART00000085298
|
l2hgdh
|
L-2-hydroxyglutarate dehydrogenase |
| chr8_+_20108592 | 1.04 |
|
|
|
| chr8_+_26015350 | 1.04 |
ENSDART00000004521
|
arih2
|
ariadne homolog 2 (Drosophila) |
| chr9_-_32347822 | 1.04 |
ENSDART00000037182
|
ankrd44
|
ankyrin repeat domain 44 |
| chr12_+_38633026 | 1.04 |
ENSDART00000155563
|
abca5
|
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr25_-_12316228 | 1.04 |
ENSDART00000168275
|
det1
|
de-etiolated homolog 1 (Arabidopsis) |
| chr19_-_46500888 | 1.03 |
ENSDART00000178772
|
ptdss1b
|
phosphatidylserine synthase 1b |
| chr8_-_17755104 | 1.03 |
ENSDART00000131276
|
si:ch211-150o23.2
|
si:ch211-150o23.2 |
| chr12_+_18794467 | 1.03 |
ENSDART00000127536
|
cbx7b
|
chromobox homolog 7b |
| chr21_-_9689702 | 1.03 |
ENSDART00000158836
|
arhgap24
|
Rho GTPase activating protein 24 |
| chr2_-_4191540 | 1.03 |
ENSDART00000158335
|
rab18b
|
RAB18B, member RAS oncogene family |
| chr21_-_30247820 | 1.02 |
ENSDART00000066363
|
ENSDARG00000045127
|
ENSDARG00000045127 |
| chr13_+_24549249 | 1.02 |
ENSDART00000113981
|
zgc:66426
|
zgc:66426 |
| chr7_-_31669675 | 1.01 |
ENSDART00000131009
|
bdnf
|
brain-derived neurotrophic factor |
| chr16_-_25318260 | 1.00 |
ENSDART00000058943
|
ENSDARG00000040280
|
ENSDARG00000040280 |
| chr19_+_7839888 | 1.00 |
|
|
|
| chr17_-_9806413 | 0.99 |
ENSDART00000021942
|
eapp
|
e2f-associated phosphoprotein |
| chr10_-_24350114 | 0.98 |
ENSDART00000109549
ENSDART00000148480 |
inpp5kb
|
inositol polyphosphate-5-phosphatase Kb |
| chr24_-_26224123 | 0.98 |
ENSDART00000080039
|
lrrc31
|
leucine rich repeat containing 31 |
| chr5_+_61678410 | 0.98 |
ENSDART00000074117
|
aspa
|
aspartoacylase |
| chr3_+_13780013 | 0.97 |
ENSDART00000164179
|
syce2
|
synaptonemal complex central element protein 2 |
| chr24_-_6649190 | 0.97 |
ENSDART00000042478
|
enkur
|
enkurin, TRPC channel interacting protein |
| chr6_-_19556029 | 0.97 |
ENSDART00000136019
|
ppp1r12c
|
protein phosphatase 1, regulatory subunit 12C |
| chr20_+_34485425 | 0.97 |
ENSDART00000146924
|
trmt1l
|
tRNA methyltransferase 1-like |
| chr13_-_22772577 | 0.97 |
ENSDART00000089133
|
rufy2
|
RUN and FYVE domain containing 2 |
| chr8_-_25696852 | 0.97 |
ENSDART00000007482
|
tspy
|
testis specific protein, Y-linked |
| chr7_-_31346844 | 0.96 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr23_+_38313746 | 0.96 |
ENSDART00000129593
|
znf217
|
zinc finger protein 217 |
| chr13_+_36832090 | 0.96 |
ENSDART00000024386
|
frmd6
|
FERM domain containing 6 |
| chr1_+_21244242 | 0.96 |
ENSDART00000087729
|
kdm4c
|
lysine (K)-specific demethylase 4C |
| chr6_-_49802414 | 0.96 |
ENSDART00000008959
|
nelfcd
|
negative elongation factor complex member C/D |
| chr8_-_50899562 | 0.96 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long-chain family member 2 |
| chr15_+_27000467 | 0.96 |
ENSDART00000023842
|
ppm1da
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Da |
| chr7_+_52437748 | 0.95 |
ENSDART00000174061
ENSDART00000174094 ENSDART00000110906 ENSDART00000174238 ENSDART00000174071 |
znf280d
|
zinc finger protein 280D |
| chr23_-_30837726 | 0.95 |
ENSDART00000075918
|
pcmtd2
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
| chr21_+_15505169 | 0.95 |
ENSDART00000011318
|
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr15_+_19948916 | 0.95 |
ENSDART00000054416
|
dyrk1ab
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, b |
| chr20_+_27813565 | 0.95 |
ENSDART00000008306
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr21_-_32048130 | 0.95 |
ENSDART00000137878
|
mat2b
|
methionine adenosyltransferase II, beta |
| chr21_-_30247560 | 0.95 |
ENSDART00000066363
|
ENSDARG00000045127
|
ENSDARG00000045127 |
| chr23_+_28213261 | 0.95 |
ENSDART00000143362
|
BX663515.1
|
ENSDARG00000094837 |
| chr14_+_7596207 | 0.94 |
ENSDART00000113299
|
zgc:110843
|
zgc:110843 |
| chr5_-_31889551 | 0.94 |
ENSDART00000168870
|
gpr107
|
G protein-coupled receptor 107 |
| chr24_-_6649248 | 0.94 |
ENSDART00000042478
|
enkur
|
enkurin, TRPC channel interacting protein |
| chr10_-_8839096 | 0.94 |
ENSDART00000080763
|
si:dkey-27b3.2
|
si:dkey-27b3.2 |
| chr2_+_34128846 | 0.94 |
ENSDART00000056666
|
klhl20
|
kelch-like family member 20 |
| chr10_-_24350023 | 0.93 |
ENSDART00000109549
ENSDART00000148480 |
inpp5kb
|
inositol polyphosphate-5-phosphatase Kb |
| chr15_+_19948869 | 0.93 |
ENSDART00000054416
|
dyrk1ab
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, b |
| chr19_+_40750511 | 0.93 |
ENSDART00000147391
|
fam133b
|
family with sequence similarity 133, member B |
| chr17_-_23234771 | 0.92 |
|
|
|
| chr14_+_27811681 | 0.91 |
|
|
|
| chr16_-_37518912 | 0.91 |
|
|
|
| chr2_-_24413154 | 0.91 |
ENSDART00000145526
|
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr4_+_11203919 | 0.91 |
ENSDART00000150551
|
BX784029.3
|
ENSDARG00000096218 |
| chr19_-_10996809 | 0.91 |
ENSDART00000163179
|
pip5k1aa
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, a |
| chr5_-_48028931 | 0.91 |
ENSDART00000031194
|
lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr23_-_18851683 | 0.91 |
ENSDART00000169180
ENSDART00000006482 |
trpc4apb
|
transient receptor potential cation channel, subfamily C, member 4 associated protein b |
| chr23_+_28213293 | 0.91 |
ENSDART00000143362
|
BX663515.1
|
ENSDARG00000094837 |
| chr20_-_223222 | 0.91 |
ENSDART00000104790
|
znf292b
|
zinc finger protein 292b |
| chr15_-_17088606 | 0.90 |
ENSDART00000154719
ENSDART00000014465 |
hip1
|
huntingtin interacting protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.3 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.6 | 2.8 | GO:0060149 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.5 | 2.7 | GO:0070874 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.5 | 5.7 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.5 | 2.0 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.5 | 1.5 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.5 | 3.7 | GO:0070266 | necroptotic process(GO:0070266) protein linear deubiquitination(GO:1990108) |
| 0.4 | 2.2 | GO:1904869 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.4 | 1.2 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.4 | 2.5 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.3 | 1.9 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.3 | 1.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.3 | 2.0 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.3 | 0.9 | GO:0035092 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.3 | 1.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.3 | 2.5 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.3 | 0.8 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.3 | 1.0 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.3 | 0.8 | GO:0070841 | inclusion body assembly(GO:0070841) regulation of inclusion body assembly(GO:0090083) |
| 0.2 | 1.9 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 | 1.1 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.2 | 2.4 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.2 | 1.1 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.2 | 0.6 | GO:0043476 | pigment accumulation(GO:0043476) |
| 0.2 | 1.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.2 | 1.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 5.4 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.2 | 0.5 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.2 | 1.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 0.5 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) hepatic duct development(GO:0061011) |
| 0.2 | 0.9 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.2 | 0.5 | GO:0042126 | nitrate metabolic process(GO:0042126) nitrate assimilation(GO:0042128) |
| 0.1 | 0.7 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.1 | 1.0 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.3 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.1 | 1.1 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.1 | 2.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 0.6 | GO:0045190 | B cell activation involved in immune response(GO:0002312) isotype switching(GO:0045190) |
| 0.1 | 0.6 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.5 | GO:0050746 | regulation of lipoprotein metabolic process(GO:0050746) regulation of protein lipidation(GO:1903059) |
| 0.1 | 1.6 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 1.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 6.5 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.1 | 1.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 1.6 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 | 0.9 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 1.3 | GO:0007584 | response to nutrient(GO:0007584) |
| 0.1 | 1.6 | GO:0090169 | regulation of spindle assembly(GO:0090169) regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 0.7 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 2.1 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 1.4 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.5 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.1 | 0.9 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 1.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 0.7 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 3.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 1.0 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.4 | GO:0006498 | N-terminal protein lipidation(GO:0006498) N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.1 | 0.6 | GO:0009651 | hypotonic response(GO:0006971) response to salt stress(GO:0009651) hypotonic salinity response(GO:0042539) sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 4.0 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 0.5 | GO:0035794 | positive regulation of mitochondrial membrane permeability(GO:0035794) mitochondrial outer membrane permeabilization(GO:0097345) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.1 | 0.6 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 0.7 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.6 | GO:1900045 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.7 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.5 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 2.1 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 0.4 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 8.9 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.1 | 1.4 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 0.5 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.5 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.9 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 0.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 1.5 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 0.7 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.1 | 0.9 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.3 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 1.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 1.3 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 | 0.3 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 | 0.4 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 3.2 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 2.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 1.0 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.1 | 0.7 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.3 | GO:0031061 | bulbus arteriosus development(GO:0003232) negative regulation of histone methylation(GO:0031061) |
| 0.1 | 1.7 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.1 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.6 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 1.5 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.1 | GO:0035494 | regulation of synaptic vesicle priming(GO:0010807) SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.7 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.4 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 2.1 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.9 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 1.7 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.2 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 1.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.2 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.6 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 2.8 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.6 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.3 | GO:0050670 | regulation of mononuclear cell proliferation(GO:0032944) regulation of T cell proliferation(GO:0042129) regulation of lymphocyte proliferation(GO:0050670) regulation of leukocyte proliferation(GO:0070663) |
| 0.0 | 0.6 | GO:0051307 | meiotic chromosome separation(GO:0051307) |
| 0.0 | 1.2 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.4 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 2.0 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 0.8 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.8 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 1.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.0 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 1.3 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 1.4 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 2.4 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.0 | 2.6 | GO:0032870 | cellular response to hormone stimulus(GO:0032870) |
| 0.0 | 0.8 | GO:0006282 | regulation of DNA repair(GO:0006282) |
| 0.0 | 0.1 | GO:0035188 | hatching(GO:0035188) hatching gland development(GO:0048785) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.9 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.7 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.2 | GO:0019856 | pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.6 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.5 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 0.5 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 0.8 | GO:0048916 | posterior lateral line development(GO:0048916) |
| 0.0 | 0.9 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.3 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 1.2 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.3 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.7 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.6 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.6 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.4 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.5 | GO:0007281 | germ cell development(GO:0007281) |
| 0.0 | 0.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.1 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) positive regulation of mRNA processing(GO:0050685) |
| 0.0 | 0.1 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.9 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0000801 | central element(GO:0000801) |
| 0.6 | 3.8 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.4 | 1.2 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.3 | 2.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.3 | 1.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.2 | 1.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.2 | 1.6 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 0.9 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 0.9 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.2 | 1.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 2.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 5.9 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 2.1 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 0.6 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 0.7 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.4 | GO:0034990 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 2.8 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 3.3 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 0.8 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.0 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 1.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.7 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.3 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 1.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 2.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 2.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.0 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 0.7 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 1.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.5 | GO:0072379 | ER membrane insertion complex(GO:0072379) |
| 0.1 | 1.0 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.6 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 2.7 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 0.7 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 3.4 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 1.2 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.6 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.1 | 0.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 1.0 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 1.0 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 3.2 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 1.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.5 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 3.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 1.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.1 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 1.7 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 7.0 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 4.1 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.8 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.5 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 4.1 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.4 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 0.2 | GO:0000784 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 1.1 | 3.2 | GO:0097020 | COPII adaptor activity(GO:0097020) |
| 0.8 | 2.5 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.8 | 2.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.7 | 2.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.7 | 2.7 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.6 | 2.3 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.5 | 1.6 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.5 | 3.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.5 | 5.6 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.4 | 1.2 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.4 | 1.5 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.4 | 1.5 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.4 | 4.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.4 | 1.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.3 | 3.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.3 | 1.9 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 0.9 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.3 | 2.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.3 | 0.9 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.3 | 0.8 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.3 | 0.8 | GO:0048270 | methionine adenosyltransferase regulator activity(GO:0048270) |
| 0.2 | 0.7 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.2 | 1.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 2.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 0.7 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.2 | 2.4 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.2 | 1.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.2 | 0.6 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 1.0 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.2 | 1.4 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.2 | 1.0 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.2 | 1.4 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.2 | 1.6 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.2 | 0.6 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.2 | 0.6 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) cGMP binding(GO:0030553) |
| 0.2 | 1.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 0.7 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.2 | 0.5 | GO:0046857 | nitrite reductase [NAD(P)H] activity(GO:0008942) oxidoreductase activity, acting on other nitrogenous compounds as donors, with NAD or NADP as acceptor(GO:0046857) |
| 0.2 | 0.5 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 0.7 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.6 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 0.4 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 1.6 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.9 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 2.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.7 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.9 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 1.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.4 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.1 | 1.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.6 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.1 | GO:0043142 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.1 | 3.0 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.4 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.7 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.9 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.2 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 1.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 1.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.6 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 1.1 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 1.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.7 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.9 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 0.7 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.1 | 0.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.6 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.4 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.2 | GO:0046624 | ceramide transporter activity(GO:0035620) sphingolipid transporter activity(GO:0046624) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 0.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.8 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.1 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 1.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 1.3 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.0 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 7.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 4.2 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 2.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.8 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 1.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 2.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 1.9 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 2.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.9 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.8 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.8 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.8 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 2.3 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.0 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 1.0 | GO:0042803 | protein homodimerization activity(GO:0042803) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 3.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 3.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 0.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.0 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.2 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.2 | 1.8 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 4.7 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.8 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 2.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 2.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 2.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 0.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.6 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 1.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.7 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.7 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 5.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.3 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.9 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.1 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |