Project
DANIO-CODE
Navigation
Downloads
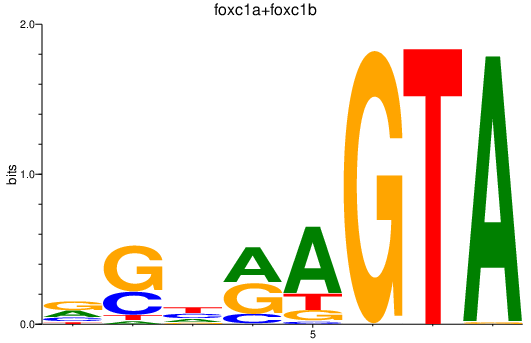
Results for foxc1a+foxc1b
Z-value: 0.83
Transcription factors associated with foxc1a+foxc1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxc1b
|
ENSDARG00000055398 | forkhead box C1b |
|
foxc1a
|
ENSDARG00000091481 | forkhead box C1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxc1a | dr10_dc_chr2_-_816669_816714 | -0.79 | 2.9e-04 | Click! |
| foxc1b | dr10_dc_chr20_+_26803282_26803322 | -0.72 | 1.6e-03 | Click! |
Activity profile of foxc1a+foxc1b motif
Sorted Z-values of foxc1a+foxc1b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of foxc1a+foxc1b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_24567184 | 2.99 |
ENSDART00000131530
|
fam113
|
family with sequence similarity 113 |
| chr11_+_41777220 | 2.35 |
ENSDART00000172846
|
camta1
|
calmodulin binding transcription activator 1 |
| chr21_-_2296253 | 2.28 |
ENSDART00000162867
|
zgc:66483
|
zgc:66483 |
| chr14_+_29945327 | 2.17 |
ENSDART00000173090
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr14_-_41101660 | 1.75 |
ENSDART00000003170
|
mid1ip1l
|
MID1 interacting protein 1, like |
| chr19_+_40558066 | 1.73 |
ENSDART00000049968
|
si:ch211-173p18.3
|
si:ch211-173p18.3 |
| chr12_+_22448972 | 1.65 |
|
|
|
| chr11_-_18090243 | 1.44 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr7_-_24567002 | 1.40 |
ENSDART00000139455
|
fam113
|
family with sequence similarity 113 |
| chr12_-_27121226 | 1.31 |
ENSDART00000153101
|
psme3
|
proteasome activator subunit 3 |
| chr12_+_29121368 | 1.25 |
ENSDART00000006505
|
mxtx2
|
mix-type homeobox gene 2 |
| chr5_-_56953587 | 1.25 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr13_-_28286577 | 1.23 |
ENSDART00000140989
|
BX950184.1
|
ENSDARG00000093030 |
| chr5_-_62303298 | 1.22 |
ENSDART00000047143
|
specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr21_+_43183522 | 1.20 |
ENSDART00000151350
|
aff4
|
AF4/FMR2 family, member 4 |
| chr20_-_28531019 | 1.19 |
ENSDART00000172133
|
CABZ01057122.1
|
ENSDARG00000105192 |
| chr10_+_33629943 | 1.19 |
ENSDART00000130093
|
c10h21orf59
|
c10h21orf59 homolog (H. sapiens) |
| chr18_-_12889296 | 1.09 |
ENSDART00000130343
|
parp12a
|
poly (ADP-ribose) polymerase family, member 12a |
| chr5_-_56953716 | 1.09 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr22_-_3661783 | 1.09 |
ENSDART00000153634
|
FP565432.1
|
ENSDARG00000097145 |
| chr12_-_27121343 | 1.06 |
ENSDART00000002835
|
psme3
|
proteasome activator subunit 3 |
| chr19_-_5186692 | 1.06 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr11_+_17849608 | 1.06 |
ENSDART00000020283
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr6_-_44164161 | 1.05 |
ENSDART00000035513
|
shq1
|
SHQ1, H/ACA ribonucleoprotein assembly factor |
| chr5_-_12086676 | 0.98 |
ENSDART00000081494
ENSDART00000162780 |
vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr17_+_45489354 | 0.97 |
ENSDART00000103493
|
ENSDARG00000070487
|
ENSDARG00000070487 |
| chr8_-_26703757 | 0.96 |
ENSDART00000148033
|
kazna
|
kazrin, periplakin interacting protein a |
| chr5_-_18393623 | 0.93 |
ENSDART00000010101
ENSDART00000099434 |
aacs
|
acetoacetyl-CoA synthetase |
| chr9_+_1820601 | 0.93 |
|
|
|
| chr1_-_11191824 | 0.92 |
ENSDART00000163971
ENSDART00000123431 |
iqce
|
IQ motif containing E |
| chr3_+_33608070 | 0.91 |
|
|
|
| chr6_+_21887963 | 0.91 |
ENSDART00000157796
|
cbx8b
|
chromobox homolog 8b |
| chr18_-_12889369 | 0.90 |
ENSDART00000130343
|
parp12a
|
poly (ADP-ribose) polymerase family, member 12a |
| chr24_-_36382704 | 0.85 |
ENSDART00000154858
|
si:ch211-40k21.5
|
si:ch211-40k21.5 |
| chr21_+_20350218 | 0.85 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr16_+_41065840 | 0.84 |
ENSDART00000124543
|
dek
|
DEK proto-oncogene |
| chr7_+_69290790 | 0.83 |
ENSDART00000109507
|
RAP1GDS1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr1_-_44649941 | 0.81 |
ENSDART00000110390
|
ENSDARG00000025518
|
ENSDARG00000025518 |
| chr3_+_5125139 | 0.81 |
ENSDART00000161755
|
CU681842.1
|
ENSDARG00000102795 |
| chr3_-_20912934 | 0.80 |
ENSDART00000156275
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr23_+_35870640 | 0.80 |
ENSDART00000139057
|
BX465864.2
|
ENSDARG00000095755 |
| chr12_+_22449611 | 0.79 |
|
|
|
| chr17_-_18777398 | 0.77 |
ENSDART00000045991
|
vrk1
|
vaccinia related kinase 1 |
| chr14_+_7596207 | 0.72 |
ENSDART00000113299
|
zgc:110843
|
zgc:110843 |
| chr15_-_6969345 | 0.71 |
ENSDART00000169529
|
mrps22
|
mitochondrial ribosomal protein S22 |
| chr21_-_18238558 | 0.70 |
ENSDART00000126672
|
brd3a
|
bromodomain containing 3a |
| chr10_-_302984 | 0.69 |
ENSDART00000157582
|
emsy
|
EMSY BRCA2-interacting transcriptional repressor |
| chr24_-_17001295 | 0.69 |
|
|
|
| chr6_+_40925259 | 0.68 |
ENSDART00000002728
ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr3_-_7709509 | 0.66 |
|
|
|
| chr16_-_39245082 | 0.65 |
ENSDART00000058546
|
ebag9
|
estrogen receptor binding site associated, antigen, 9 |
| chr25_-_34596670 | 0.65 |
ENSDART00000171659
|
zgc:162611
|
zgc:162611 |
| chr14_+_14302479 | 0.63 |
ENSDART00000165099
|
fut11
|
fucosyltransferase 11 (alpha (1,3) fucosyltransferase) |
| chr8_+_52458939 | 0.63 |
|
|
|
| chr1_+_51118765 | 0.61 |
ENSDART00000156349
|
BX548032.3
|
ENSDARG00000096092 |
| chr21_+_20350334 | 0.60 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr9_+_24381641 | 0.60 |
ENSDART00000003482
|
stk17b
|
serine/threonine kinase 17b (apoptosis-inducing) |
| chr2_-_9729223 | 0.60 |
ENSDART00000004398
|
cope
|
coatomer protein complex, subunit epsilon |
| chr22_+_26736632 | 0.59 |
|
|
|
| chr3_+_33608669 | 0.58 |
|
|
|
| chr13_-_36554590 | 0.57 |
ENSDART00000085298
|
l2hgdh
|
L-2-hydroxyglutarate dehydrogenase |
| chr25_-_4407821 | 0.57 |
|
|
|
| chr14_+_29945396 | 0.57 |
ENSDART00000173090
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr3_+_33608622 | 0.56 |
|
|
|
| chr14_-_40951773 | 0.55 |
|
|
|
| chr1_-_11191863 | 0.55 |
ENSDART00000163971
ENSDART00000123431 |
iqce
|
IQ motif containing E |
| chr24_-_55232 | 0.55 |
|
|
|
| chr5_+_31259894 | 0.54 |
ENSDART00000036235
|
iscub
|
iron-sulfur cluster assembly enzyme b |
| chr23_+_44470297 | 0.54 |
ENSDART00000149842
|
MEPCE
|
methylphosphate capping enzyme |
| chr8_+_12913443 | 0.52 |
ENSDART00000081601
|
cept1a
|
choline/ethanolamine phosphotransferase 1a |
| chr6_+_21887935 | 0.51 |
ENSDART00000157796
|
cbx8b
|
chromobox homolog 8b |
| chr6_-_51771558 | 0.51 |
ENSDART00000073847
|
blcap
|
bladder cancer associated protein |
| chr5_-_18393705 | 0.49 |
ENSDART00000010101
ENSDART00000099434 |
aacs
|
acetoacetyl-CoA synthetase |
| chr5_-_12086593 | 0.49 |
ENSDART00000081494
ENSDART00000162780 |
vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr16_+_25096821 | 0.49 |
ENSDART00000157312
|
znf1035
|
zinc finger protein 1035 |
| chr3_-_17990895 | 0.48 |
ENSDART00000175098
ENSDART00000177044 |
CU682780.3
|
ENSDARG00000107147 |
| chr5_+_38117847 | 0.48 |
|
|
|
| chr17_+_45421503 | 0.48 |
ENSDART00000147557
|
tagapa
|
T-cell activation RhoGTPase activating protein a |
| chr5_+_55448865 | 0.47 |
ENSDART00000083137
|
mrm1
|
mitochondrial rRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr12_+_26447489 | 0.47 |
ENSDART00000152955
|
si:dkey-57h18.1
|
si:dkey-57h18.1 |
| chr14_+_33723918 | 0.47 |
ENSDART00000015670
|
med7
|
mediator complex subunit 7 |
| chr5_-_66614185 | 0.46 |
ENSDART00000147053
|
rsrc2
|
arginine/serine-rich coiled-coil 2 |
| chr15_+_15520846 | 0.46 |
ENSDART00000155708
|
zgc:174895
|
zgc:174895 |
| chr7_-_19274727 | 0.46 |
ENSDART00000114203
|
man2b2
|
mannosidase, alpha, class 2B, member 2 |
| chr2_-_50391964 | 0.45 |
ENSDART00000009347
|
exoc3
|
exocyst complex component 3 |
| chr2_+_25517887 | 0.44 |
ENSDART00000110922
|
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr16_+_25097165 | 0.44 |
ENSDART00000157312
|
znf1035
|
zinc finger protein 1035 |
| chr7_+_17656099 | 0.43 |
ENSDART00000077113
|
mta2
|
metastasis associated 1 family, member 2 |
| chr16_+_41065107 | 0.43 |
ENSDART00000058586
|
dek
|
DEK proto-oncogene |
| chr17_+_45421543 | 0.42 |
ENSDART00000147557
|
tagapa
|
T-cell activation RhoGTPase activating protein a |
| chr20_+_48631844 | 0.41 |
ENSDART00000108884
|
prdm1c
|
PR domain containing 1c, with ZNF domain |
| chr23_-_45106985 | 0.41 |
ENSDART00000076373
|
st8sia7.1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 7.1 |
| chr6_-_15364606 | 0.40 |
ENSDART00000156439
|
st6gal2b
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2b |
| chr2_-_37419017 | 0.38 |
ENSDART00000015723
|
prkci
|
protein kinase C, iota |
| chr5_+_55448837 | 0.38 |
ENSDART00000083137
|
mrm1
|
mitochondrial rRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr12_+_10405904 | 0.37 |
ENSDART00000029133
|
snu13b
|
SNU13 homolog, small nuclear ribonucleoprotein b (U4/U6.U5) |
| chr6_+_56224631 | 0.37 |
ENSDART00000174612
ENSDART00000177824 |
FO834918.1
|
ENSDARG00000106015 |
| chr1_-_17544731 | 0.37 |
ENSDART00000143157
|
AL929210.1
|
ENSDARG00000094943 |
| chr10_-_302923 | 0.37 |
ENSDART00000157582
|
emsy
|
EMSY BRCA2-interacting transcriptional repressor |
| chr2_+_56016077 | 0.36 |
ENSDART00000164741
|
pgpep1
|
pyroglutamyl-peptidase I |
| chr10_+_22066527 | 0.36 |
|
|
|
| chr18_+_39079744 | 0.35 |
ENSDART00000098729
|
zgc:171509
|
zgc:171509 |
| chr6_+_33947023 | 0.35 |
ENSDART00000157397
|
orc1
|
origin recognition complex, subunit 1 |
| chr9_+_24891584 | 0.35 |
|
|
|
| chr4_-_71644701 | 0.35 |
ENSDART00000174178
|
si:cabz01021430.2
|
si:cabz01021430.2 |
| chr9_+_53810607 | 0.34 |
|
|
|
| chr22_-_7887401 | 0.34 |
ENSDART00000097198
|
sc:d217
|
sc:d217 |
| chr20_+_38555185 | 0.34 |
ENSDART00000020153
|
adck3
|
aarF domain containing kinase 3 |
| chr21_+_18238176 | 0.33 |
ENSDART00000144322
|
wdr5
|
WD repeat domain 5 |
| chr25_-_8583608 | 0.32 |
ENSDART00000159409
|
unc45a
|
unc-45 myosin chaperone A |
| chr8_+_52544578 | 0.31 |
ENSDART00000127729
|
stambpb
|
STAM binding protein b |
| chr5_+_58327436 | 0.31 |
ENSDART00000062175
|
derl2
|
derlin 2 |
| chr4_-_18976343 | 0.31 |
|
|
|
| chr21_-_27335013 | 0.31 |
|
|
|
| chr21_-_33196706 | 0.31 |
ENSDART00000003983
|
rbm22
|
RNA binding motif protein 22 |
| chr13_+_46637827 | 0.31 |
ENSDART00000167931
|
mtrf1l
|
mitochondrial translational release factor 1-like |
| chr20_+_36909452 | 0.31 |
ENSDART00000062931
|
abracl
|
ABRA C-terminal like |
| chr5_-_66614227 | 0.30 |
ENSDART00000147053
|
rsrc2
|
arginine/serine-rich coiled-coil 2 |
| chr8_-_10978942 | 0.30 |
ENSDART00000064066
|
bcas2
|
BCAS2, pre-mRNA processing factor |
| chr9_+_9956221 | 0.30 |
ENSDART00000168559
|
ttf2
|
transcription termination factor, RNA polymerase II |
| chr1_-_28146457 | 0.29 |
ENSDART00000110270
ENSDART00000170797 |
pwp2h
|
PWP2 periodic tryptophan protein homolog (yeast) |
| chr9_+_24381412 | 0.29 |
ENSDART00000003482
|
stk17b
|
serine/threonine kinase 17b (apoptosis-inducing) |
| chr3_-_40834641 | 0.28 |
ENSDART00000018676
|
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr12_+_4651511 | 0.28 |
ENSDART00000128145
|
kansl1a
|
KAT8 regulatory NSL complex subunit 1a |
| chr8_-_47339092 | 0.28 |
ENSDART00000060853
|
pex10
|
peroxisomal biogenesis factor 10 |
| chr5_-_61134500 | 0.28 |
ENSDART00000079855
|
im:7138535
|
im:7138535 |
| chr7_+_52984549 | 0.27 |
ENSDART00000169830
|
trip4
|
thyroid hormone receptor interactor 4 |
| chr5_-_1220217 | 0.27 |
ENSDART00000154344
|
ciz1a
|
cdkn1a interacting zinc finger protein 1a |
| chr10_+_24721139 | 0.26 |
ENSDART00000144710
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr15_+_1928102 | 0.26 |
ENSDART00000150104
|
sbds
|
SBDS, ribosome maturation factor |
| chr10_+_22066689 | 0.24 |
|
|
|
| chr17_+_1080921 | 0.24 |
|
|
|
| chr7_+_16854387 | 0.23 |
ENSDART00000013409
|
prmt3
|
protein arginine methyltransferase 3 |
| chr21_+_34942142 | 0.23 |
ENSDART00000139635
|
rbm11
|
RNA binding motif protein 11 |
| chr4_-_11054766 | 0.23 |
ENSDART00000161470
|
mettl25
|
methyltransferase like 25 |
| chr25_+_29245574 | 0.23 |
ENSDART00000073478
|
brd1b
|
bromodomain containing 1b |
| chr6_-_33946853 | 0.23 |
ENSDART00000057290
|
prpf38a
|
pre-mRNA processing factor 38A |
| KN150339v1_-_47655 | 0.22 |
|
|
|
| chr5_+_35839198 | 0.21 |
ENSDART00000102973
ENSDART00000103020 |
eda
|
ectodysplasin A |
| chr18_-_17796903 | 0.21 |
|
|
|
| chr5_-_55448596 | 0.21 |
ENSDART00000083134
|
mks1
|
Meckel syndrome, type 1 |
| chr11_+_19208363 | 0.20 |
ENSDART00000165906
|
prickle2b
|
prickle homolog 2b |
| chr6_+_33946990 | 0.19 |
ENSDART00000157397
|
orc1
|
origin recognition complex, subunit 1 |
| chr7_+_19230972 | 0.19 |
ENSDART00000077868
|
si:ch211-212k18.7
|
si:ch211-212k18.7 |
| chr19_-_23665476 | 0.19 |
ENSDART00000140665
|
grb10a
|
growth factor receptor-bound protein 10a |
| chr21_-_45833636 | 0.18 |
ENSDART00000161716
|
larp1
|
La ribonucleoprotein domain family, member 1 |
| chr17_-_18777348 | 0.18 |
ENSDART00000045991
|
vrk1
|
vaccinia related kinase 1 |
| chr9_+_6277952 | 0.18 |
ENSDART00000078525
|
uxs1
|
UDP-glucuronate decarboxylase 1 |
| chr2_-_37555425 | 0.17 |
ENSDART00000143496
|
arhgef18a
|
rho/rac guanine nucleotide exchange factor (GEF) 18a |
| chr1_+_54270060 | 0.17 |
ENSDART00000152528
|
r3hcc1l
|
R3H domain and coiled-coil containing 1-like |
| chr12_-_31619559 | 0.17 |
|
|
|
| chr17_-_33463210 | 0.15 |
ENSDART00000135218
|
ccdc28a
|
coiled-coil domain containing 28A |
| chr13_+_8272812 | 0.14 |
ENSDART00000091097
|
socs5b
|
suppressor of cytokine signaling 5b |
| chr16_-_31393328 | 0.14 |
ENSDART00000178298
|
mroh1
|
maestro heat-like repeat family member 1 |
| chr11_+_44696399 | 0.14 |
ENSDART00000158188
|
mgea5l
|
meningioma expressed antigen 5 (hyaluronidase) like |
| chr10_+_33630200 | 0.13 |
ENSDART00000130093
|
c10h21orf59
|
c10h21orf59 homolog (H. sapiens) |
| chr23_-_14460492 | 0.11 |
ENSDART00000019620
|
ddx23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr21_+_43183570 | 0.10 |
ENSDART00000151512
|
aff4
|
AF4/FMR2 family, member 4 |
| chr17_+_25269767 | 0.10 |
|
|
|
| chr15_-_1928150 | 0.10 |
|
|
|
| chr10_+_22066361 | 0.10 |
|
|
|
| chr22_-_10098297 | 0.09 |
ENSDART00000013835
|
bloc1s1
|
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr16_+_25096883 | 0.08 |
ENSDART00000157312
|
znf1035
|
zinc finger protein 1035 |
| chr16_-_13927684 | 0.08 |
ENSDART00000146719
|
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr13_-_9737104 | 0.08 |
ENSDART00000158381
|
myom2b
|
myomesin 2b |
| chr13_-_50299643 | 0.08 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr2_+_20655215 | 0.08 |
|
|
|
| chr15_+_36355348 | 0.08 |
|
|
|
| chr16_-_13927947 | 0.08 |
ENSDART00000144856
|
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr20_+_31372117 | 0.07 |
|
|
|
| chr11_+_14046768 | 0.06 |
ENSDART00000059752
|
med16
|
mediator complex subunit 16 |
| chr15_-_20189405 | 0.06 |
ENSDART00000152355
|
med13b
|
mediator complex subunit 13b |
| chr7_-_16345978 | 0.05 |
ENSDART00000128488
|
e2f8
|
E2F transcription factor 8 |
| chr2_+_37854303 | 0.05 |
ENSDART00000143203
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr16_+_5625788 | 0.05 |
|
|
|
| chr16_+_5625456 | 0.05 |
|
|
|
| chr23_+_22858773 | 0.04 |
ENSDART00000142085
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr10_+_22065010 | 0.03 |
ENSDART00000133304
|
npm1a
|
nucleophosmin 1a (nucleolar phosphoprotein B23, numatrin) |
| chr8_-_51380776 | 0.02 |
ENSDART00000060628
|
chmp7
|
charged multivesicular body protein 7 |
| chr24_-_37912374 | 0.02 |
|
|
|
| chr4_-_71644771 | 0.02 |
ENSDART00000174198
|
si:cabz01021430.2
|
si:cabz01021430.2 |
| chr7_-_34142109 | 0.01 |
ENSDART00000174546
|
madd
|
MAP-kinase activating death domain |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0090277 | positive regulation of peptide secretion(GO:0002793) positive regulation of insulin secretion(GO:0032024) positive regulation of hormone secretion(GO:0046887) positive regulation of peptide hormone secretion(GO:0090277) |
| 0.4 | 1.1 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.3 | 1.0 | GO:0000493 | box H/ACA snoRNP assembly(GO:0000493) |
| 0.3 | 0.9 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.2 | 1.4 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 0.5 | GO:0040014 | regulation of multicellular organism growth(GO:0040014) |
| 0.1 | 1.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 1.1 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.4 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.1 | 2.4 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.1 | 0.6 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.2 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 1.7 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.0 | 1.3 | GO:2000779 | regulation of double-strand break repair(GO:2000779) |
| 0.0 | 0.7 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.3 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.5 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 1.1 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.3 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.2 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.7 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.2 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 1.3 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.5 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.1 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.6 | GO:0000974 | Prp19 complex(GO:0000974) U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.3 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.7 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.7 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.3 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.7 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0031730 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) CCR5 chemokine receptor binding(GO:0031730) |
| 0.5 | 2.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.4 | 1.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.2 | 0.6 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 0.5 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 0.3 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 1.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 2.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.4 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.1 | 1.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.4 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.5 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.5 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.4 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 2.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.9 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 2.7 | GO:0008017 | microtubule binding(GO:0008017) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.4 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.6 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |