Project
DANIO-CODE
Navigation
Downloads
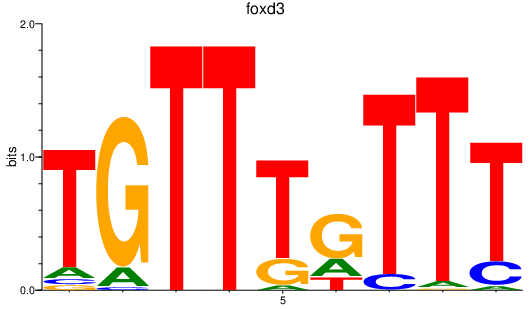
Results for foxd3
Z-value: 1.97
Transcription factors associated with foxd3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxd3
|
ENSDARG00000021032 | forkhead box D3 |
Activity profile of foxd3 motif
Sorted Z-values of foxd3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of foxd3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_32914487 | 5.44 |
ENSDART00000015547
|
cldng
|
claudin g |
| chr5_+_36168475 | 5.13 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr15_+_38397772 | 5.03 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr5_+_57254393 | 5.00 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr17_+_14957568 | 4.17 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr4_-_16639774 | 3.97 |
ENSDART00000125323
|
caprin2
|
caprin family member 2 |
| chr12_-_28680035 | 3.73 |
ENSDART00000020667
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr10_+_15644868 | 3.60 |
ENSDART00000139259
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr7_-_47978449 | 3.58 |
ENSDART00000127007
ENSDART00000024062 |
cpeb1b
|
cytoplasmic polyadenylation element binding protein 1b |
| chr7_-_47990610 | 3.52 |
ENSDART00000147968
|
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr1_-_7160294 | 3.50 |
ENSDART00000174972
|
BX908756.2
|
ENSDARG00000108483 |
| chr22_-_28828375 | 3.46 |
ENSDART00000104880
ENSDART00000005112 |
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr12_+_28774063 | 3.44 |
ENSDART00000166229
|
phkg2
|
phosphorylase kinase, gamma 2 (testis) |
| chr24_+_36429525 | 3.33 |
ENSDART00000062722
|
pus3
|
pseudouridylate synthase 3 |
| chr1_-_50395003 | 3.21 |
ENSDART00000035150
|
spast
|
spastin |
| chr12_-_22116694 | 3.19 |
ENSDART00000038310
ENSDART00000132731 |
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr9_+_55731780 | 3.16 |
ENSDART00000111111
|
nlgn4b
|
neuroligin 4b |
| chr11_+_17849608 | 3.15 |
ENSDART00000020283
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr2_+_30013086 | 3.08 |
ENSDART00000138050
|
rbm33b
|
RNA binding motif protein 33b |
| chr7_-_51497945 | 3.07 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr15_+_20054306 | 3.06 |
ENSDART00000155199
|
zgc:112083
|
zgc:112083 |
| chr22_+_24532560 | 3.00 |
ENSDART00000169847
|
wdr47b
|
WD repeat domain 47b |
| chr16_-_47446494 | 2.92 |
ENSDART00000032188
|
si:dkey-256h2.1
|
si:dkey-256h2.1 |
| chr19_-_27093095 | 2.88 |
|
|
|
| chr10_-_14971273 | 2.84 |
ENSDART00000137430
|
smad2
|
SMAD family member 2 |
| chr8_+_37716781 | 2.80 |
ENSDART00000108556
|
npm2a
|
nucleophosmin/nucleoplasmin, 2a |
| chr22_-_10562118 | 2.78 |
ENSDART00000105846
|
si:dkey-42i9.8
|
si:dkey-42i9.8 |
| chr2_-_44924505 | 2.76 |
ENSDART00000113351
|
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr16_+_25344257 | 2.74 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| chr1_-_7160099 | 2.70 |
ENSDART00000174972
|
BX908756.2
|
ENSDARG00000108483 |
| chr18_+_2250457 | 2.67 |
|
|
|
| chr10_+_19059739 | 2.67 |
ENSDART00000038674
|
tmem230a
|
transmembrane protein 230a |
| chr4_+_14983045 | 2.65 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr4_-_20414881 | 2.65 |
ENSDART00000161343
|
cep83
|
centrosomal protein 83 |
| chr5_-_8712114 | 2.64 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr20_+_48631807 | 2.62 |
ENSDART00000108884
|
prdm1c
|
PR domain containing 1c, with ZNF domain |
| chr19_+_5399535 | 2.57 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr3_+_17562348 | 2.57 |
|
|
|
| chr22_+_24532479 | 2.57 |
ENSDART00000169847
|
wdr47b
|
WD repeat domain 47b |
| chr16_+_47283253 | 2.48 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr20_+_13987091 | 2.46 |
ENSDART00000152611
|
nek2
|
NIMA-related kinase 2 |
| chr2_-_39026651 | 2.45 |
ENSDART00000114085
|
si:ch211-119o8.6
|
si:ch211-119o8.6 |
| chr19_-_6274169 | 2.44 |
ENSDART00000140347
ENSDART00000092656 |
erf
|
Ets2 repressor factor |
| chr2_+_22984662 | 2.43 |
ENSDART00000144890
|
bokb
|
BCL2-related ovarian killer b |
| chr15_-_17163526 | 2.43 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr17_+_23463072 | 2.42 |
|
|
|
| chr18_+_15303434 | 2.39 |
ENSDART00000099777
ENSDART00000170246 |
si:dkey-103i16.6
|
si:dkey-103i16.6 |
| chr23_+_25952724 | 2.36 |
ENSDART00000124963
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr3_-_34398743 | 2.34 |
ENSDART00000023039
|
sept9a
|
septin 9a |
| KN150339v1_-_39357 | 2.34 |
ENSDART00000169638
|
wu:fa19b12
|
wu:fa19b12 |
| chr6_-_53049946 | 2.33 |
ENSDART00000103267
|
fam212ab
|
family with sequence similarity 212, member Ab |
| chr22_+_18840143 | 2.33 |
|
|
|
| chr19_+_16159980 | 2.31 |
ENSDART00000151169
|
gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr12_+_18784456 | 2.30 |
ENSDART00000105854
|
josd1
|
Josephin domain containing 1 |
| chr5_+_60677040 | 2.30 |
ENSDART00000009507
|
gatsl2
|
GATS protein-like 2 |
| chr2_+_34984631 | 2.29 |
ENSDART00000141796
|
astn1
|
astrotactin 1 |
| chr10_+_37457234 | 2.26 |
ENSDART00000136554
|
akap1a
|
A kinase (PRKA) anchor protein 1a |
| chr16_+_25693740 | 2.26 |
ENSDART00000077484
|
zhx2a
|
zinc fingers and homeoboxes 2a |
| chr16_+_36794614 | 2.25 |
ENSDART00000139069
|
decr1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr24_+_9887575 | 2.25 |
ENSDART00000172773
|
si:ch211-146l10.8
|
si:ch211-146l10.8 |
| chr15_+_21776031 | 2.23 |
ENSDART00000136151
|
NKAPD1
|
zgc:162339 |
| chr15_+_38397715 | 2.22 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr16_+_19831573 | 2.22 |
ENSDART00000135359
|
macc1
|
metastasis associated in colon cancer 1 |
| chr20_-_6542402 | 2.21 |
ENSDART00000054653
|
mcm3l
|
MCM3 minichromosome maintenance deficient 3 (S. cerevisiae), like |
| chr19_-_48346883 | 2.21 |
ENSDART00000123771
ENSDART00000081675 |
ctnnb2
|
catenin, beta 2 |
| KN150663v1_-_3381 | 2.20 |
|
|
|
| chr11_-_10001506 | 2.19 |
ENSDART00000123359
|
nlgn1
|
neuroligin 1 |
| chr4_+_1770595 | 2.19 |
ENSDART00000148486
|
scaf11
|
SR-related CTD-associated factor 11 |
| chr5_-_15971338 | 2.19 |
ENSDART00000110437
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr18_+_17545564 | 2.18 |
ENSDART00000061007
|
mt2
|
metallothionein 2 |
| chr24_-_20225190 | 2.17 |
ENSDART00000109223
|
dlec1
|
deleted in lung and esophageal cancer 1 |
| chr5_-_8712068 | 2.13 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr20_-_28897936 | 2.13 |
ENSDART00000049462
|
rab15
|
RAB15, member RAS oncogene family |
| chr8_+_8605240 | 2.12 |
ENSDART00000075624
|
usp11
|
ubiquitin specific peptidase 11 |
| chr12_+_30389706 | 2.10 |
|
|
|
| chr15_-_14102102 | 2.09 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr17_-_23689233 | 2.09 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr19_-_35408237 | 2.07 |
ENSDART00000141704
|
si:rp71-45k5.2
|
si:rp71-45k5.2 |
| chr15_-_43996427 | 2.05 |
ENSDART00000177294
ENSDART00000177892 ENSDART00000174655 |
FO704909.1
|
ENSDARG00000106999 |
| chr3_-_29779725 | 2.05 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr2_-_40857927 | 2.05 |
ENSDART00000157099
|
CT583650.1
|
ENSDARG00000096869 |
| chr23_+_45177338 | 2.04 |
|
|
|
| chr8_+_40463218 | 2.01 |
|
|
|
| chr14_-_32467474 | 2.00 |
|
|
|
| chr6_+_40925259 | 2.00 |
ENSDART00000002728
ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr6_-_53145464 | 1.97 |
ENSDART00000079694
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr23_-_35384257 | 1.96 |
ENSDART00000113643
|
fbxo25
|
F-box protein 25 |
| chr16_+_25693878 | 1.96 |
ENSDART00000077484
|
zhx2a
|
zinc fingers and homeoboxes 2a |
| chr19_+_29716509 | 1.96 |
ENSDART00000142694
ENSDART00000009149 |
tmem57a
|
transmembrane protein 57a |
| chr3_-_28371648 | 1.95 |
ENSDART00000150912
|
CR792426.1
|
ENSDARG00000096347 |
| chr9_-_51745361 | 1.94 |
ENSDART00000154959
|
AL954669.1
|
ENSDARG00000097096 |
| chr12_+_20569459 | 1.92 |
ENSDART00000112847
|
st6galnac1.2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1, tandem duplicate 2 |
| chr5_-_47419494 | 1.90 |
ENSDART00000078401
|
tmem161b
|
transmembrane protein 161B |
| chr15_-_8787051 | 1.89 |
ENSDART00000154171
|
arhgap35a
|
Rho GTPase activating protein 35a |
| chr19_-_41820114 | 1.88 |
ENSDART00000038038
|
shfm1
|
split hand/foot malformation (ectrodactyly) type 1 |
| chr3_-_29779598 | 1.87 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr16_-_41537827 | 1.86 |
ENSDART00000169312
|
cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr9_-_8916860 | 1.83 |
ENSDART00000138527
|
rab20
|
RAB20, member RAS oncogene family |
| chr23_+_25952913 | 1.81 |
ENSDART00000124963
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr19_+_29716557 | 1.80 |
ENSDART00000142694
ENSDART00000009149 |
tmem57a
|
transmembrane protein 57a |
| chr5_-_21523599 | 1.80 |
ENSDART00000023306
|
asb12a
|
ankyrin repeat and SOCS box-containing 12a |
| chr5_-_22466262 | 1.79 |
ENSDART00000172549
|
si:dkey-237j10.2
|
si:dkey-237j10.2 |
| chr14_-_7437682 | 1.79 |
ENSDART00000172215
|
si:ch211-39i2.2
|
si:ch211-39i2.2 |
| chr3_+_30187036 | 1.79 |
ENSDART00000151006
|
CR936968.1
|
ENSDARG00000096295 |
| chr12_-_10438255 | 1.78 |
ENSDART00000106172
|
rac1a
|
ras-related C3 botulinum toxin substrate 1a (rho family, small GTP binding protein Rac1) |
| chr1_+_32825950 | 1.76 |
ENSDART00000018472
|
chmp2bb
|
charged multivesicular body protein 2Bb |
| chr14_-_34293596 | 1.75 |
ENSDART00000128869
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr22_-_10722533 | 1.75 |
ENSDART00000081191
|
sass6
|
SAS-6 centriolar assembly protein |
| chr1_+_50547385 | 1.74 |
ENSDART00000132141
|
btbd3a
|
BTB (POZ) domain containing 3a |
| chr20_-_28898117 | 1.73 |
ENSDART00000049462
|
rab15
|
RAB15, member RAS oncogene family |
| chr15_+_23721725 | 1.72 |
ENSDART00000078336
|
klc3
|
kinesin light chain 3 |
| chr6_-_39220951 | 1.72 |
ENSDART00000133305
ENSDART00000166290 |
os9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr24_+_12873142 | 1.71 |
ENSDART00000142457
|
stau2
|
staufen double-stranded RNA binding protein 2 |
| chr12_-_17741377 | 1.71 |
ENSDART00000166604
|
baiap2l1a
|
BAI1-associated protein 2-like 1a |
| chr3_-_42949449 | 1.71 |
ENSDART00000161127
|
axin1
|
axin 1 |
| chr5_+_12971972 | 1.70 |
ENSDART00000132406
|
cnnm4b
|
cyclin and CBS domain divalent metal cation transport mediator 4b |
| chr13_+_8364704 | 1.70 |
ENSDART00000109059
|
ttc7a
|
tetratricopeptide repeat domain 7A |
| chr12_-_25288550 | 1.69 |
ENSDART00000142674
|
zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr8_-_602656 | 1.69 |
|
|
|
| chr11_+_24756894 | 1.68 |
ENSDART00000129211
|
ENSDARG00000044874
|
ENSDARG00000044874 |
| chr2_-_56897037 | 1.68 |
ENSDART00000164086
|
slc25a42
|
solute carrier family 25, member 42 |
| chr9_-_38213786 | 1.67 |
|
|
|
| chr18_+_27095471 | 1.67 |
ENSDART00000125326
ENSDART00000098334 |
ppp1r15b
|
protein phosphatase 1, regulatory subunit 15B |
| chr18_+_47305642 | 1.67 |
ENSDART00000009775
|
rbm7
|
RNA binding motif protein 7 |
| chr7_-_49528331 | 1.66 |
ENSDART00000052083
|
fjx1
|
four jointed box 1 |
| chr16_+_47283374 | 1.65 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr20_+_39145226 | 1.65 |
ENSDART00000142778
|
BX569784.1
|
ENSDARG00000092980 |
| chr17_+_32670368 | 1.64 |
ENSDART00000142449
|
ctsba
|
cathepsin Ba |
| chr16_-_26654610 | 1.62 |
ENSDART00000134448
|
l3mbtl1b
|
l(3)mbt-like 1b (Drosophila) |
| chr5_-_56254482 | 1.62 |
ENSDART00000014028
|
ppm1db
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Db |
| chr14_-_26167494 | 1.62 |
ENSDART00000143454
ENSDART00000111860 |
syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr17_-_29253770 | 1.62 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr23_+_25952958 | 1.62 |
ENSDART00000124963
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr1_+_45148002 | 1.61 |
ENSDART00000148086
|
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr15_+_38397897 | 1.60 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr25_+_33787063 | 1.59 |
|
|
|
| chr2_+_44659334 | 1.59 |
ENSDART00000155017
|
pask
|
PAS domain containing serine/threonine kinase |
| chr19_-_6274458 | 1.57 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr17_-_7661725 | 1.57 |
ENSDART00000143870
|
rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr17_-_29253918 | 1.56 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr18_+_38963423 | 1.55 |
|
|
|
| chr2_+_35134340 | 1.55 |
ENSDART00000113609
|
rfwd2
|
ring finger and WD repeat domain 2 |
| chr4_+_9466175 | 1.54 |
ENSDART00000012659
|
zgc:55888
|
zgc:55888 |
| chr3_-_42949526 | 1.53 |
ENSDART00000161127
|
axin1
|
axin 1 |
| chr7_-_58856806 | 1.53 |
ENSDART00000159285
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr23_-_33811895 | 1.53 |
ENSDART00000131680
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr3_-_26060787 | 1.52 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr10_-_6587281 | 1.52 |
ENSDART00000163788
ENSDART00000171833 |
chd1
|
chromodomain helicase DNA binding protein 1 |
| chr19_-_9584480 | 1.51 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr14_-_36096780 | 1.51 |
ENSDART00000173350
|
aga
|
aspartylglucosaminidase |
| chr20_-_33584483 | 1.51 |
ENSDART00000177645
|
smek1
|
SMEK homolog 1, suppressor of mek1 (Dictyostelium) |
| chr24_-_12685892 | 1.51 |
ENSDART00000039312
|
ipo4
|
importin 4 |
| chr8_-_4270732 | 1.50 |
ENSDART00000134378
|
cux2b
|
cut-like homeobox 2b |
| chr6_-_51811024 | 1.50 |
ENSDART00000031597
|
psmf1
|
proteasome inhibitor subunit 1 |
| chr13_+_18376540 | 1.50 |
ENSDART00000142622
|
si:ch211-198a12.6
|
si:ch211-198a12.6 |
| chr8_+_39734109 | 1.49 |
ENSDART00000017153
|
hps4
|
Hermansky-Pudlak syndrome 4 |
| chr9_-_12603178 | 1.49 |
|
|
|
| chr8_+_17107665 | 1.49 |
ENSDART00000158707
ENSDART00000061758 |
mier3b
|
mesoderm induction early response 1, family member 3 b |
| chr5_+_29052293 | 1.49 |
ENSDART00000035400
|
tsc1a
|
tuberous sclerosis 1a |
| chr6_-_52567422 | 1.49 |
ENSDART00000098421
|
uqcc1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr2_+_1645259 | 1.48 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr5_+_3683209 | 1.48 |
ENSDART00000132056
|
ggnbp2
|
gametogenetin binding protein 2 |
| chr10_+_29251234 | 1.47 |
ENSDART00000123033
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr2_+_30013537 | 1.47 |
ENSDART00000151848
|
rbm33b
|
RNA binding motif protein 33b |
| chr5_-_23013803 | 1.46 |
ENSDART00000137655
|
stag2a
|
stromal antigen 2a |
| chr21_+_5854755 | 1.46 |
ENSDART00000149689
|
mob1bb
|
MOB kinase activator 1Bb |
| chr19_+_24483983 | 1.46 |
ENSDART00000141351
|
pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr3_+_40142679 | 1.44 |
ENSDART00000009411
|
bud31
|
BUD31 homolog (S. cerevisiae) |
| chr17_-_7661762 | 1.43 |
ENSDART00000135538
|
rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr14_-_36096895 | 1.42 |
ENSDART00000173350
|
aga
|
aspartylglucosaminidase |
| chr16_+_46443800 | 1.42 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr19_-_43359719 | 1.42 |
ENSDART00000086659
|
cyhr1
|
cysteine/histidine-rich 1 |
| chr10_+_14530451 | 1.41 |
ENSDART00000026383
|
sigmar1
|
sigma non-opioid intracellular receptor 1 |
| chr19_-_6274237 | 1.41 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr10_-_33307992 | 1.41 |
ENSDART00000169064
|
bcl7ba
|
B-cell CLL/lymphoma 7B, a |
| chr12_+_20290592 | 1.41 |
ENSDART00000115015
|
arhgap17a
|
Rho GTPase activating protein 17a |
| chr6_-_31377513 | 1.40 |
ENSDART00000145715
|
ak4
|
adenylate kinase 4 |
| chr21_+_15608741 | 1.40 |
ENSDART00000146909
|
chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr5_-_14000166 | 1.39 |
ENSDART00000099566
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr4_+_1727494 | 1.39 |
ENSDART00000149448
|
AL929171.1
|
ENSDARG00000095920 |
| chr13_-_22831005 | 1.39 |
ENSDART00000143112
|
tspan15
|
tetraspanin 15 |
| chr17_+_15780112 | 1.38 |
ENSDART00000027667
ENSDART00000161637 |
rragd
|
ras-related GTP binding D |
| chr17_-_29254409 | 1.38 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr3_-_56803756 | 1.37 |
ENSDART00000014103
|
zgc:112148
|
zgc:112148 |
| chr7_+_28824986 | 1.37 |
ENSDART00000052349
|
ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr23_-_33753484 | 1.37 |
ENSDART00000138416
|
tfcp2
|
transcription factor CP2 |
| chr3_-_42949479 | 1.35 |
ENSDART00000161127
|
axin1
|
axin 1 |
| chr9_+_8919663 | 1.35 |
ENSDART00000134954
|
carkd
|
carbohydrate kinase domain containing |
| chr23_+_36608246 | 1.35 |
ENSDART00000113179
|
tspan31
|
tetraspanin 31 |
| chr15_+_35089305 | 1.34 |
ENSDART00000156515
|
zgc:55621
|
zgc:55621 |
| chr1_-_54488824 | 1.34 |
ENSDART00000150430
|
pane1
|
proliferation associated nuclear element |
| chr1_-_54505313 | 1.34 |
ENSDART00000100619
ENSDART00000163796 |
LUC7L2
|
zgc:158803 |
| chr17_+_37563386 | 1.34 |
ENSDART00000156281
|
mta1
|
metastasis associated 1 |
| chr5_-_47419466 | 1.33 |
ENSDART00000078401
|
tmem161b
|
transmembrane protein 161B |
| chr18_+_20571460 | 1.33 |
ENSDART00000151990
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr22_-_17663766 | 1.33 |
ENSDART00000147070
|
tjp3
|
tight junction protein 3 |
| chr5_-_47419401 | 1.33 |
ENSDART00000078401
|
tmem161b
|
transmembrane protein 161B |
| chr19_-_42933592 | 1.33 |
ENSDART00000007642
|
zgc:110239
|
zgc:110239 |
| chr12_-_28680442 | 1.33 |
ENSDART00000178777
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr18_-_44854242 | 1.32 |
ENSDART00000086823
|
srpr
|
signal recognition particle receptor (docking protein) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 7.3 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 1.0 | 4.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 1.0 | 4.0 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 1.0 | 2.9 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 1.0 | 6.7 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.9 | 2.7 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.9 | 0.9 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.9 | 3.6 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.9 | 3.5 | GO:0051150 | regulation of smooth muscle cell differentiation(GO:0051150) negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.8 | 3.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) mRNA pseudouridine synthesis(GO:1990481) |
| 0.8 | 2.5 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.8 | 3.2 | GO:0043243 | positive regulation of protein complex disassembly(GO:0043243) |
| 0.8 | 7.6 | GO:0060281 | regulation of oocyte development(GO:0060281) |
| 0.7 | 2.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.7 | 2.2 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.7 | 2.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.6 | 3.9 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.6 | 3.2 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.6 | 3.7 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.6 | 2.9 | GO:0010922 | positive regulation of phosphatase activity(GO:0010922) positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.6 | 5.7 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.6 | 1.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.6 | 1.7 | GO:0019376 | galactosylceramide catabolic process(GO:0006683) galactolipid catabolic process(GO:0019376) |
| 0.5 | 2.6 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.5 | 1.5 | GO:0002504 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.5 | 3.0 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.5 | 1.5 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.5 | 2.9 | GO:0050427 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.5 | 1.9 | GO:0015882 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.5 | 6.3 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.5 | 2.7 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.4 | 3.5 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.4 | 1.7 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.4 | 0.4 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.4 | 4.2 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.4 | 1.5 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.4 | 1.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.4 | 1.1 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.4 | 2.8 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.3 | 1.0 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.3 | 4.1 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.3 | 4.8 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.3 | 3.0 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.3 | 1.0 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.3 | 1.9 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.3 | 1.9 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.3 | 1.6 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.3 | 1.6 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.3 | 2.2 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.3 | 2.5 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.3 | 2.2 | GO:0043536 | positive regulation of blood vessel endothelial cell migration(GO:0043536) positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.3 | 0.9 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.3 | 2.3 | GO:1902743 | regulation of lamellipodium assembly(GO:0010591) regulation of lamellipodium organization(GO:1902743) |
| 0.3 | 0.9 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.3 | 1.7 | GO:0032527 | protein exit from endoplasmic reticulum(GO:0032527) endoplasmic reticulum to cytosol transport(GO:1903513) |
| 0.3 | 3.7 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.3 | 1.7 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) |
| 0.3 | 1.6 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.3 | 2.9 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.2 | 1.5 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 1.0 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.2 | 6.3 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.2 | 1.5 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 1.2 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.2 | 0.7 | GO:0090281 | negative regulation of vascular permeability(GO:0043116) negative regulation of calcium ion import(GO:0090281) |
| 0.2 | 1.4 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.2 | 1.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.2 | 0.7 | GO:0042126 | nitrate metabolic process(GO:0042126) nitrate assimilation(GO:0042128) |
| 0.2 | 0.5 | GO:0071921 | regulation of sister chromatid cohesion(GO:0007063) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.2 | 1.3 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.2 | 0.7 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 1.3 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.2 | 2.2 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.2 | 1.1 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.2 | 4.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.2 | 1.5 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.2 | 0.4 | GO:0003179 | heart valve morphogenesis(GO:0003179) |
| 0.2 | 0.6 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.2 | 3.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.2 | 0.6 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.2 | 2.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 0.6 | GO:0032570 | response to progesterone(GO:0032570) medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.2 | 2.6 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.2 | 0.8 | GO:0046078 | dUMP biosynthetic process(GO:0006226) dUMP metabolic process(GO:0046078) |
| 0.2 | 0.6 | GO:0006408 | snRNA export from nucleus(GO:0006408) regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.2 | 2.7 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.2 | 1.5 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.2 | 0.4 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) |
| 0.2 | 2.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 0.6 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.2 | 0.9 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.2 | 0.7 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 2.0 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.2 | 0.7 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 1.6 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.2 | 0.5 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.2 | 2.1 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.2 | 1.0 | GO:0033198 | response to ATP(GO:0033198) |
| 0.2 | 0.7 | GO:2001271 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 0.7 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.2 | 0.6 | GO:0048103 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 0.2 | 1.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 0.9 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.2 | 4.8 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.2 | 0.8 | GO:0034367 | macromolecular complex remodeling(GO:0034367) |
| 0.1 | 4.0 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 0.6 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 1.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.9 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.4 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.1 | 0.4 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 2.4 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.1 | 1.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.5 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 1.2 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.1 | 0.7 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.1 | 0.9 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 3.3 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.1 | 1.8 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 1.6 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 0.4 | GO:0002686 | negative regulation of leukocyte migration(GO:0002686) |
| 0.1 | 3.0 | GO:2001236 | regulation of extrinsic apoptotic signaling pathway(GO:2001236) |
| 0.1 | 1.6 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.3 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 2.2 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 0.6 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 1.0 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.1 | 1.2 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 1.6 | GO:0033866 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.1 | 0.4 | GO:0045190 | B cell activation involved in immune response(GO:0002312) isotype switching(GO:0045190) |
| 0.1 | 0.5 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.4 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.1 | 1.2 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 0.8 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.1 | 0.3 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 1.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.3 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.1 | 1.9 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 0.3 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) |
| 0.1 | 0.7 | GO:0071169 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 1.5 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 1.3 | GO:0008105 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.1 | 0.9 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.4 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.1 | 0.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.5 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.9 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 1.1 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.1 | 1.2 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.4 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 0.4 | GO:0000423 | macromitophagy(GO:0000423) |
| 0.1 | 2.0 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 0.8 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 0.3 | GO:0051580 | neurotransmitter uptake(GO:0001504) regulation of neurotransmitter uptake(GO:0051580) neurotransmitter reuptake(GO:0098810) |
| 0.1 | 4.0 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 0.2 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.1 | 0.2 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.1 | 1.3 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 1.5 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 0.7 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 2.3 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.1 | 1.3 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.1 | 0.8 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 2.4 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 4.3 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 0.7 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.1 | 0.4 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.8 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 0.7 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.8 | GO:0035794 | positive regulation of mitochondrial membrane permeability(GO:0035794) mitochondrial outer membrane permeabilization(GO:0097345) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.1 | 2.4 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.1 | 0.4 | GO:0090316 | positive regulation of intracellular protein transport(GO:0090316) |
| 0.1 | 0.4 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.4 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.1 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 1.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.7 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.1 | 1.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 0.8 | GO:2000380 | regulation of mesoderm development(GO:2000380) |
| 0.1 | 1.7 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.1 | 0.3 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.1 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.1 | 2.0 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 1.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.3 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 0.1 | GO:1903429 | regulation of cell maturation(GO:1903429) |
| 0.0 | 0.7 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.1 | GO:0055016 | hypochord development(GO:0055016) |
| 0.0 | 2.7 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 1.0 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 2.1 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 2.9 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 1.2 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 2.1 | GO:0022406 | membrane docking(GO:0022406) vesicle docking(GO:0048278) |
| 0.0 | 0.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 4.0 | GO:1990778 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 0.7 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.8 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 1.4 | GO:0009306 | protein secretion(GO:0009306) |
| 0.0 | 2.9 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.8 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.7 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.0 | 1.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.8 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 14.3 | GO:0051603 | proteolysis involved in cellular protein catabolic process(GO:0051603) |
| 0.0 | 1.7 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 1.0 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 3.4 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.2 | GO:0009084 | glutamine family amino acid biosynthetic process(GO:0009084) |
| 0.0 | 0.6 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:1901031 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.6 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.3 | GO:0009113 | purine nucleobase biosynthetic process(GO:0009113) |
| 0.0 | 0.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.6 | GO:0007164 | establishment of planar polarity(GO:0001736) establishment of tissue polarity(GO:0007164) |
| 0.0 | 0.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 2.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.3 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.4 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.5 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 2.0 | GO:0017148 | negative regulation of translation(GO:0017148) negative regulation of cellular amide metabolic process(GO:0034249) |
| 0.0 | 0.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 5.8 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.0 | 3.3 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 1.2 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.4 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.7 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 1.4 | GO:0001666 | response to hypoxia(GO:0001666) response to decreased oxygen levels(GO:0036293) |
| 0.0 | 0.2 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 0.8 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 0.2 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 | 0.7 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.3 | GO:0072666 | protein targeting to vacuole(GO:0006623) establishment of protein localization to vacuole(GO:0072666) |
| 0.0 | 0.2 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.0 | 0.3 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.3 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.4 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.3 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.4 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 0.4 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 2.0 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.9 | 3.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.8 | 3.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.7 | 7.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.7 | 2.7 | GO:0098536 | deuterosome(GO:0098536) |
| 0.5 | 3.6 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.5 | 5.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.5 | 1.5 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.5 | 3.9 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.4 | 1.2 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.3 | 3.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.3 | 2.9 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.3 | 2.2 | GO:0071546 | pi-body(GO:0071546) |
| 0.3 | 2.7 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.3 | 4.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.3 | 3.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.3 | 1.2 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.3 | 0.6 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.3 | 3.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.3 | 1.0 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.3 | 2.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.3 | 4.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.3 | 2.0 | GO:0089701 | U2AF(GO:0089701) |
| 0.2 | 0.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.2 | 0.7 | GO:1990745 | EARP complex(GO:1990745) |
| 0.2 | 1.6 | GO:0070187 | telosome(GO:0070187) |
| 0.2 | 1.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.2 | 4.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 1.8 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 1.0 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.2 | 0.6 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 1.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 2.5 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.2 | 9.1 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.2 | 6.5 | GO:0030496 | midbody(GO:0030496) |
| 0.2 | 0.7 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.2 | 1.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 9.3 | GO:0005814 | centriole(GO:0005814) |
| 0.2 | 1.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 4.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 0.9 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 2.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 2.0 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 1.0 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.1 | 0.7 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 1.9 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 3.2 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 7.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.3 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.1 | 0.3 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.1 | 1.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 3.6 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 0.7 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 1.4 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.2 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.6 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 4.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 5.8 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 16.6 | GO:0005764 | lysosome(GO:0005764) |
| 0.1 | 1.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.3 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.1 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.5 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 0.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 2.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 2.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 10.7 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 1.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 4.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 2.2 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.6 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 1.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.2 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.4 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.6 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.1 | GO:0032021 | NELF complex(GO:0032021) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.7 | 3.7 | GO:0003948 | N4-(beta-N-acetylglucosaminyl)-L-asparaginase activity(GO:0003948) |
| 0.7 | 2.0 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.6 | 6.5 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.6 | 3.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.6 | 1.7 | GO:0004336 | galactosylceramidase activity(GO:0004336) |
| 0.5 | 1.6 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.5 | 2.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.5 | 1.6 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.5 | 3.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.5 | 2.0 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.5 | 0.5 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.5 | 6.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.5 | 2.9 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.4 | 3.8 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.4 | 1.9 | GO:0070551 | endoribonuclease activity, cleaving siRNA-paired mRNA(GO:0070551) |
| 0.4 | 1.9 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.4 | 1.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.4 | 1.1 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.3 | 1.0 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.3 | 1.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.3 | 2.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.3 | 1.9 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.3 | 2.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.3 | 2.4 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.3 | 1.5 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.3 | 4.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.3 | 4.1 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.3 | 0.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.3 | 1.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.3 | 1.6 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.3 | 4.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.3 | 5.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.2 | 1.0 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.2 | 1.9 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 1.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.2 | 0.7 | GO:0008942 | nitrite reductase [NAD(P)H] activity(GO:0008942) oxidoreductase activity, acting on other nitrogenous compounds as donors, with NAD or NADP as acceptor(GO:0046857) |
| 0.2 | 2.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 1.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 2.0 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.2 | 0.7 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 0.9 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 0.7 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.2 | 0.6 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.2 | 3.6 | GO:0004407 | histone deacetylase activity(GO:0004407) NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.2 | 5.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.2 | 1.0 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 0.4 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.2 | 0.9 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.2 | 2.0 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.2 | 4.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 1.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 1.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 1.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.2 | 1.0 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.2 | 1.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.7 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.1 | 2.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 1.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 3.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 6.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.8 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.1 | 1.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.9 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 1.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.5 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 0.5 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 2.5 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 1.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.9 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 0.3 | GO:0004044 | amidophosphoribosyltransferase activity(GO:0004044) |
| 0.1 | 1.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 3.2 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.1 | 0.6 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 1.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.5 | GO:0070273 | phosphatidylinositol-5-phosphate binding(GO:0010314) phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 1.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 2.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.9 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 1.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.2 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.1 | 0.9 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 1.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 1.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 1.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.7 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 11.1 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 11.0 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.1 | 1.3 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 1.7 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.1 | 2.9 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.1 | 2.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.4 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.3 | GO:0001097 | TFIIH-class transcription factor binding(GO:0001097) poly(C) RNA binding(GO:0017130) |
| 0.0 | 1.9 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 15.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 3.7 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 2.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 1.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 2.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.7 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.8 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 1.1 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.8 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.7 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.6 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.2 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 3.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.5 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 7.5 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 1.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.0 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.0 | 0.3 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.3 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 2.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 2.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.0 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 3.5 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 9.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.3 | 4.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 6.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 1.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 1.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 6.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 2.8 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.1 | 3.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 1.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 2.3 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 1.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 3.3 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 1.9 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 3.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 1.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 1.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 1.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 1.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.7 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.5 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.4 | 7.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 4.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 3.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 0.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.2 | 1.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 10.1 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.2 | 1.7 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.2 | 0.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 2.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 2.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.5 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.1 | 0.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 5.4 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.1 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 1.0 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 0.9 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 1.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 1.7 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 2.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.9 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 0.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.6 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 1.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.5 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 2.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.2 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.1 | 0.5 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 0.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.9 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |
| 0.0 | 2.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 0.0 | 2.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 0.3 | REACTOME METABOLISM OF RNA | Genes involved in Metabolism of RNA |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |