Project
DANIO-CODE
Navigation
Downloads
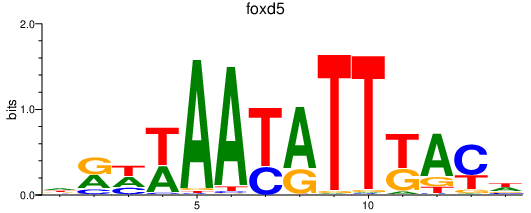
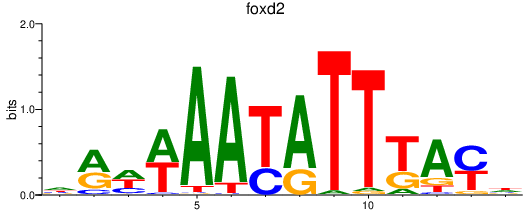
Results for foxd5_foxd2
Z-value: 1.10
Transcription factors associated with foxd5_foxd2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxd5
|
ENSDARG00000042485 | forkhead box D5 |
|
foxd2
|
ENSDARG00000058133 | forkhead box D2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxd2 | dr10_dc_chr8_+_19642272_19642279 | 0.60 | 1.5e-02 | Click! |
| foxd5 | dr10_dc_chr8_+_30443651_30443762 | 0.30 | 2.7e-01 | Click! |
Activity profile of foxd5_foxd2 motif
Sorted Z-values of foxd5_foxd2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of foxd5_foxd2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_17570306 | 3.90 |
ENSDART00000139361
|
gpx4a
|
glutathione peroxidase 4a |
| chr1_-_29669484 | 3.37 |
ENSDART00000164202
|
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr19_-_22802777 | 2.89 |
ENSDART00000151234
|
eppk1
|
epiplakin 1 |
| chr4_+_12014166 | 2.64 |
ENSDART00000034401
ENSDART00000172691 |
cry1aa
|
cryptochrome circadian clock 1aa |
| chr18_-_46356213 | 2.52 |
ENSDART00000010813
|
foxa3
|
forkhead box A3 |
| chr1_-_29669529 | 2.46 |
ENSDART00000164202
|
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr4_-_25226680 | 2.28 |
ENSDART00000066932
ENSDART00000066933 ENSDART00000157714 |
itih2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr17_-_10682357 | 2.19 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr20_+_4120302 | 2.09 |
ENSDART00000159837
|
cnksr3
|
cnksr family member 3 |
| chr25_-_7635824 | 1.89 |
ENSDART00000126499
|
phf21ab
|
PHD finger protein 21Ab |
| chr5_-_21523520 | 1.74 |
ENSDART00000023306
|
asb12a
|
ankyrin repeat and SOCS box-containing 12a |
| chr12_-_6144233 | 1.74 |
ENSDART00000152292
|
a1cf
|
apobec1 complementation factor |
| chr3_+_36282205 | 1.73 |
ENSDART00000170318
|
si:ch1073-443f11.2
|
si:ch1073-443f11.2 |
| chr8_+_15968469 | 1.69 |
ENSDART00000134787
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr13_+_32013916 | 1.68 |
ENSDART00000020270
|
osr1
|
odd-skipped related transciption factor 1 |
| chr5_+_31606213 | 1.61 |
ENSDART00000126873
|
myhz2
|
myosin, heavy polypeptide 2, fast muscle specific |
| chr8_+_7937128 | 1.59 |
ENSDART00000137920
|
si:ch211-169p10.1
|
si:ch211-169p10.1 |
| chr17_+_21944679 | 1.56 |
ENSDART00000063704
|
crip3
|
cysteine-rich protein 3 |
| chr20_-_39003917 | 1.55 |
|
|
|
| chr2_+_27000558 | 1.45 |
ENSDART00000099208
|
asph
|
aspartate beta-hydroxylase |
| chr25_+_386916 | 1.43 |
|
|
|
| chr14_+_2969679 | 1.37 |
ENSDART00000044678
|
ENSDARG00000034011
|
ENSDARG00000034011 |
| chr25_+_19051298 | 1.35 |
ENSDART00000067324
|
mfge8b
|
milk fat globule-EGF factor 8 protein b |
| chr6_-_55881470 | 1.30 |
ENSDART00000160991
|
cyp24a1
|
cytochrome P450, family 24, subfamily A, polypeptide 1 |
| chr25_+_24152717 | 1.29 |
ENSDART00000064646
|
tmem86a
|
transmembrane protein 86A |
| chr2_-_9950224 | 1.27 |
ENSDART00000146407
|
si:ch1073-170o4.1
|
si:ch1073-170o4.1 |
| chr10_-_8336541 | 1.25 |
ENSDART00000163803
|
plpp1a
|
phospholipid phosphatase 1a |
| chr22_+_11726312 | 1.23 |
ENSDART00000155366
|
krt96
|
keratin 96 |
| chr14_-_4014025 | 1.21 |
ENSDART00000157782
|
irf2
|
interferon regulatory factor 2 |
| chr2_+_48052054 | 1.17 |
ENSDART00000159701
|
ftr23
|
finTRIM family, member 23 |
| chr3_-_21149752 | 1.16 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr20_-_39003972 | 1.15 |
|
|
|
| chr7_-_12656353 | 1.10 |
ENSDART00000023301
ENSDART00000172901 ENSDART00000163045 ENSDART00000163461 |
sh3gl3a
|
SH3-domain GRB2-like 3a |
| chr8_+_48488009 | 1.07 |
|
|
|
| chr6_-_8029184 | 1.01 |
ENSDART00000091628
|
ccdc151
|
coiled-coil domain containing 151 |
| chr25_-_7635442 | 0.96 |
ENSDART00000021577
|
phf21ab
|
PHD finger protein 21Ab |
| chr6_-_32718634 | 0.93 |
ENSDART00000175666
|
mafaa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Aa |
| chr19_-_31815128 | 0.93 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr2_+_43736542 | 0.91 |
ENSDART00000148454
|
itgb1b.1
|
integrin, beta 1b.1 |
| chr8_+_7937320 | 0.91 |
ENSDART00000137920
|
si:ch211-169p10.1
|
si:ch211-169p10.1 |
| chr9_+_4188263 | 0.90 |
ENSDART00000164543
|
kalrna
|
kalirin RhoGEF kinase a |
| chr6_+_40673803 | 0.87 |
ENSDART00000111639
|
rereb
|
arginine-glutamic acid dipeptide (RE) repeats b |
| chr14_-_24304373 | 0.87 |
|
|
|
| chr11_+_41401893 | 0.86 |
ENSDART00000171655
|
AL929185.1
|
ENSDARG00000103982 |
| chr24_+_9911748 | 0.86 |
|
|
|
| chr5_+_36049885 | 0.84 |
ENSDART00000147667
|
alkbh6
|
alkB homolog 6 |
| chr5_+_34798432 | 0.83 |
ENSDART00000098010
|
ptger4b
|
prostaglandin E receptor 4 (subtype EP4) b |
| chr16_+_11833310 | 0.82 |
ENSDART00000060266
|
ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr23_-_20332076 | 0.81 |
ENSDART00000147326
|
lamb2
|
laminin, beta 2 (laminin S) |
| chr8_-_16223730 | 0.81 |
ENSDART00000057590
|
dmrta2
|
DMRT-like family A2 |
| chr17_-_9917832 | 0.80 |
ENSDART00000161574
ENSDART00000166081 |
zgc:109986
|
zgc:109986 |
| chr22_-_11408859 | 0.77 |
ENSDART00000007649
|
mid1ip1b
|
MID1 interacting protein 1b |
| chr9_+_16437408 | 0.77 |
ENSDART00000006787
|
epha3
|
eph receptor A3 |
| chr20_+_2937165 | 0.77 |
ENSDART00000135919
|
amd1
|
adenosylmethionine decarboxylase 1 |
| chr16_-_30606364 | 0.75 |
ENSDART00000147986
|
lmna
|
lamin A |
| chr25_+_28717079 | 0.75 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase, muscle, b |
| chr22_-_13018196 | 0.74 |
ENSDART00000028787
|
ahr1b
|
aryl hydrocarbon receptor 1b |
| chr23_+_24711233 | 0.73 |
|
|
|
| chr21_-_44610563 | 0.73 |
ENSDART00000099528
|
spry4
|
sprouty homolog 4 (Drosophila) |
| chr25_-_33920256 | 0.71 |
|
|
|
| chr1_-_36356039 | 0.70 |
ENSDART00000172111
ENSDART00000160056 |
nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr10_+_9764591 | 0.69 |
ENSDART00000091780
|
rc3h2
|
ring finger and CCCH-type domains 2 |
| chr23_+_22763861 | 0.68 |
|
|
|
| chr25_+_17327279 | 0.68 |
ENSDART00000061738
|
elmo3
|
engulfment and cell motility 3 |
| chr23_-_17034625 | 0.67 |
ENSDART00000142546
|
cdc42l2
|
cell division cycle 42 like 2 |
| chr5_+_66984082 | 0.67 |
ENSDART00000137700
|
si:dkey-70b23.2
|
si:dkey-70b23.2 |
| chr18_+_9524331 | 0.66 |
ENSDART00000053125
|
sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr12_-_34786844 | 0.66 |
ENSDART00000027379
|
bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr25_-_13580057 | 0.65 |
ENSDART00000090226
|
znf319b
|
zinc finger protein 319b |
| chr17_+_37985095 | 0.65 |
ENSDART00000075941
|
plekhh1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr6_+_4091384 | 0.65 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr20_-_13728005 | 0.64 |
ENSDART00000152499
|
ezrb
|
ezrin b |
| chr9_-_37940101 | 0.63 |
ENSDART00000087663
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr15_+_28435937 | 0.62 |
ENSDART00000142298
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr16_+_46526268 | 0.61 |
ENSDART00000134734
|
rpz5
|
rapunzel 5 |
| chr2_-_37974443 | 0.60 |
ENSDART00000034595
|
cbln10
|
cerebellin 10 |
| chr7_-_18295101 | 0.59 |
ENSDART00000173969
|
rgs12a
|
regulator of G protein signaling 12a |
| chr1_-_36356001 | 0.57 |
ENSDART00000172111
ENSDART00000160056 |
nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr25_-_7635175 | 0.55 |
ENSDART00000142439
|
phf21ab
|
PHD finger protein 21Ab |
| chr23_-_35392359 | 0.54 |
ENSDART00000166167
|
fam110c
|
family with sequence similarity 110, member C |
| chr18_+_9524184 | 0.54 |
ENSDART00000053125
|
sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr18_+_36683567 | 0.54 |
ENSDART00000098972
|
capn12
|
calpain 12 |
| chr16_+_43235751 | 0.53 |
ENSDART00000065643
|
dbf4
|
DBF4 zinc finger |
| chr5_-_14990023 | 0.53 |
|
|
|
| chr25_-_13057808 | 0.53 |
ENSDART00000172571
|
smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr10_+_4114961 | 0.52 |
|
|
|
| chr6_-_40101104 | 0.52 |
ENSDART00000017402
|
ip6k2b
|
inositol hexakisphosphate kinase 2b |
| chr19_-_5416317 | 0.52 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr7_+_28896401 | 0.52 |
ENSDART00000076345
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr18_+_9524247 | 0.51 |
ENSDART00000053125
|
sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr8_+_13064944 | 0.51 |
|
|
|
| KN150670v1_-_72156 | 0.50 |
|
|
|
| chr12_-_5420963 | 0.50 |
ENSDART00000109305
|
tbc1d12b
|
TBC1 domain family, member 12b |
| chr24_-_24306469 | 0.49 |
ENSDART00000154149
|
BX323067.1
|
ENSDARG00000097984 |
| chr8_+_15968636 | 0.48 |
ENSDART00000141173
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr18_+_19430116 | 0.48 |
ENSDART00000025107
|
map2k1
|
mitogen-activated protein kinase kinase 1 |
| chr23_+_22763789 | 0.46 |
|
|
|
| chr22_-_17570275 | 0.45 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr3_-_49104540 | 0.45 |
ENSDART00000138503
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr19_+_42492029 | 0.44 |
ENSDART00000150949
|
nfyc
|
nuclear transcription factor Y, gamma |
| chr22_+_24686610 | 0.43 |
ENSDART00000088027
|
ssx2ipb
|
synovial sarcoma, X breakpoint 2 interacting protein b |
| chr14_-_26078923 | 0.43 |
ENSDART00000022236
|
emx3
|
empty spiracles homeobox 3 |
| chr24_+_9163779 | 0.43 |
|
|
|
| chr3_-_21149711 | 0.43 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr15_-_28329664 | 0.41 |
ENSDART00000175022
|
prpf8
|
pre-mRNA processing factor 8 |
| chr16_+_22950567 | 0.40 |
ENSDART00000143957
|
flad1
|
flavin adenine dinucleotide synthetase 1 |
| chr20_+_36330995 | 0.40 |
ENSDART00000131867
|
cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr22_+_2244267 | 0.40 |
ENSDART00000142890
|
si:dkey-4c15.9
|
si:dkey-4c15.9 |
| chr11_+_30416953 | 0.39 |
|
|
|
| chr5_-_11443522 | 0.39 |
ENSDART00000074979
|
rnft2
|
ring finger protein, transmembrane 2 |
| chr1_+_32964583 | 0.37 |
ENSDART00000131664
|
nsun3
|
NOP2/Sun domain family, member 3 |
| chr2_-_32404140 | 0.35 |
ENSDART00000148202
|
ubtfl
|
upstream binding transcription factor, like |
| chr10_+_39325810 | 0.35 |
ENSDART00000170079
|
foxred1
|
FAD-dependent oxidoreductase domain containing 1 |
| chr9_+_3164027 | 0.35 |
ENSDART00000160567
ENSDART00000161216 |
dync1i2a
|
dynein, cytoplasmic 1, intermediate chain 2a |
| chr22_+_11726429 | 0.35 |
ENSDART00000155366
|
krt96
|
keratin 96 |
| chr11_-_9479592 | 0.34 |
ENSDART00000175829
|
CU179643.1
|
ENSDARG00000086489 |
| chr19_+_32679258 | 0.33 |
|
|
|
| chr17_+_30431520 | 0.33 |
ENSDART00000153826
|
lpin1
|
lipin 1 |
| chr19_+_32679320 | 0.33 |
|
|
|
| chr2_+_57894141 | 0.33 |
ENSDART00000171264
ENSDART00000163278 |
si:ch211-155e24.3
|
si:ch211-155e24.3 |
| chr11_+_6298048 | 0.32 |
ENSDART00000104409
ENSDART00000139882 |
ranbp3a
|
RAN binding protein 3a |
| chr14_+_34626233 | 0.32 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr25_-_10671899 | 0.31 |
ENSDART00000121724
|
lrp5
|
low density lipoprotein receptor-related protein 5 |
| chr15_+_24756002 | 0.31 |
ENSDART00000110618
|
LRRC75A
|
leucine rich repeat containing 75A |
| chr15_+_24277664 | 0.30 |
ENSDART00000155502
|
sez6b
|
seizure related 6 homolog b |
| chr1_-_661529 | 0.30 |
ENSDART00000166786
ENSDART00000170483 |
appa
|
amyloid beta (A4) precursor protein a |
| chr6_+_28232386 | 0.29 |
ENSDART00000163057
|
FP101882.1
|
ENSDARG00000098121 |
| chr20_-_26973962 | 0.29 |
|
|
|
| chr25_+_5567388 | 0.28 |
|
|
|
| chr16_-_7310925 | 0.27 |
ENSDART00000149030
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr16_-_8711133 | 0.26 |
|
|
|
| chr6_-_43030859 | 0.26 |
ENSDART00000161722
|
glyctk
|
glycerate kinase |
| chr21_+_27379622 | 0.25 |
ENSDART00000108763
|
cfb
|
complement factor B |
| chr20_-_51454057 | 0.24 |
ENSDART00000023064
|
slc35b2
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr3_-_34207879 | 0.24 |
|
|
|
| chr6_-_10837245 | 0.24 |
ENSDART00000141017
|
sumo3a
|
small ubiquitin-like modifier 3a |
| chr23_-_28496509 | 0.24 |
ENSDART00000140448
|
c1ql4a
|
complement component 1, q subcomponent-like 4 |
| chr18_+_5005303 | 0.23 |
|
|
|
| chr5_+_29071526 | 0.23 |
ENSDART00000043096
|
ak8
|
adenylate kinase 8 |
| chr22_-_16153692 | 0.23 |
ENSDART00000171331
|
vcam1b
|
vascular cell adhesion molecule 1b |
| chr15_+_28435875 | 0.22 |
ENSDART00000168453
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr19_+_5218864 | 0.22 |
ENSDART00000108541
|
si:dkey-89b17.4
|
si:dkey-89b17.4 |
| chr6_+_4091251 | 0.22 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr5_-_22847399 | 0.21 |
ENSDART00000169258
|
nlgn3b
|
neuroligin 3b |
| chr23_+_20779487 | 0.21 |
ENSDART00000079538
|
ccdc30
|
coiled-coil domain containing 30 |
| chr5_-_13844114 | 0.21 |
|
|
|
| chr9_-_6230087 | 0.21 |
ENSDART00000032368
|
tmtops2b
|
teleost multiple tissue opsin 2b |
| chr11_-_9479689 | 0.21 |
ENSDART00000175829
|
CU179643.1
|
ENSDARG00000086489 |
| chr22_+_5633739 | 0.21 |
ENSDART00000106141
|
tma7
|
translation machinery associated 7 homolog |
| chr4_-_9866451 | 0.21 |
ENSDART00000133169
|
fbxo18
|
F-box protein, helicase, 18 |
| chr16_-_1479139 | 0.20 |
ENSDART00000036348
|
sim1a
|
single-minded family bHLH transcription factor 1a |
| chr2_-_30710221 | 0.19 |
ENSDART00000090292
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr5_+_26195283 | 0.19 |
ENSDART00000053001
|
tcn2
|
transcobalamin II |
| chr16_-_43235448 | 0.19 |
ENSDART00000165293
|
slc25a40
|
solute carrier family 25, member 40 |
| chr18_+_19430260 | 0.17 |
ENSDART00000025107
|
map2k1
|
mitogen-activated protein kinase kinase 1 |
| chr14_+_20809902 | 0.17 |
ENSDART00000139103
|
aldob
|
aldolase b, fructose-bisphosphate |
| chr18_-_6844440 | 0.17 |
ENSDART00000142647
|
si:dkey-266m15.5
|
si:dkey-266m15.5 |
| chr16_-_12282596 | 0.17 |
ENSDART00000110567
|
clstn3
|
calsyntenin 3 |
| chr5_+_36049995 | 0.16 |
ENSDART00000111414
|
alkbh6
|
alkB homolog 6 |
| chr22_-_11463487 | 0.16 |
ENSDART00000002691
|
tspan7b
|
tetraspanin 7b |
| chr6_+_24561921 | 0.14 |
ENSDART00000169283
|
znf644b
|
zinc finger protein 644b |
| chr9_+_30297924 | 0.14 |
ENSDART00000060174
|
jagn1a
|
jagunal homolog 1a |
| chr19_+_816354 | 0.14 |
ENSDART00000093304
|
nrm
|
nurim (nuclear envelope membrane protein |
| chr16_+_43236190 | 0.13 |
ENSDART00000065643
|
dbf4
|
DBF4 zinc finger |
| chr13_-_34926382 | 0.13 |
ENSDART00000178470
|
btbd3b
|
BTB (POZ) domain containing 3b |
| chr20_-_25537241 | 0.13 |
ENSDART00000153266
|
itsn2a
|
intersectin 2a |
| chr19_+_5219163 | 0.12 |
|
|
|
| chr16_+_53391438 | 0.12 |
ENSDART00000010792
|
ptdss1a
|
phosphatidylserine synthase 1a |
| chr18_-_46355957 | 0.12 |
ENSDART00000010813
|
foxa3
|
forkhead box A3 |
| chr8_+_26390972 | 0.12 |
ENSDART00000053447
|
ifrd2
|
interferon-related developmental regulator 2 |
| chr25_-_7635613 | 0.12 |
ENSDART00000021577
|
phf21ab
|
PHD finger protein 21Ab |
| chr6_-_40700033 | 0.11 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| KN150442v1_-_37820 | 0.11 |
ENSDART00000158371
|
ptmab
|
prothymosin, alpha b |
| chr20_+_34814623 | 0.10 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr20_-_49307929 | 0.10 |
ENSDART00000057700
|
naa20
|
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
| chr13_+_32609784 | 0.10 |
ENSDART00000160138
|
sobpa
|
sine oculis binding protein homolog (Drosophila) a |
| chr1_-_9256864 | 0.10 |
ENSDART00000041849
|
tmem8a
|
transmembrane protein 8A |
| chr16_+_18729942 | 0.10 |
ENSDART00000147844
ENSDART00000021596 |
rxrbb
|
retinoid x receptor, beta b |
| chr23_+_22763739 | 0.09 |
|
|
|
| chr19_-_42668754 | 0.08 |
ENSDART00000132591
|
si:ch211-191i18.4
|
si:ch211-191i18.4 |
| chr2_-_37151666 | 0.08 |
ENSDART00000146123
|
elavl1
|
ELAV like RNA binding protein 1 |
| chr3_+_45345181 | 0.07 |
|
|
|
| chr17_+_9918253 | 0.06 |
ENSDART00000169892
|
srp54
|
signal recognition particle 54 |
| chr20_-_26973934 | 0.06 |
|
|
|
| chr8_+_35483042 | 0.06 |
ENSDART00000111021
|
si:dkeyp-14d3.1
|
si:dkeyp-14d3.1 |
| chr14_-_24304628 | 0.06 |
|
|
|
| chr18_+_10957421 | 0.05 |
ENSDART00000166561
|
ttc38
|
tetratricopeptide repeat domain 38 |
| chr20_+_16822321 | 0.05 |
ENSDART00000161226
|
psmc1b
|
proteasome 26S subunit, ATPase 1b |
| chr15_-_5911548 | 0.04 |
|
|
|
| chr19_+_46631965 | 0.03 |
ENSDART00000158703
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr18_+_22296882 | 0.03 |
ENSDART00000079139
|
ctcf
|
CCCTC-binding factor (zinc finger protein) |
| chr13_+_32609849 | 0.02 |
ENSDART00000160138
|
sobpa
|
sine oculis binding protein homolog (Drosophila) a |
| chr13_+_16390948 | 0.02 |
ENSDART00000122557
|
kcnma1a
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1a |
| chr4_-_9866515 | 0.02 |
ENSDART00000133169
|
fbxo18
|
F-box protein, helicase, 18 |
| chr24_+_3447129 | 0.02 |
ENSDART00000134598
|
wdr37
|
WD repeat domain 37 |
| chr6_-_10837765 | 0.01 |
ENSDART00000143730
|
sumo3a
|
small ubiquitin-like modifier 3a |
| chr8_-_52607867 | 0.01 |
ENSDART00000176559
|
ENSDARG00000099294
|
ENSDARG00000099294 |
| chr21_+_37668334 | 0.00 |
ENSDART00000143621
|
nsd1b
|
nuclear receptor binding SET domain protein 1b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.7 | 2.6 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.5 | 1.4 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.5 | 2.9 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.5 | 1.9 | GO:0035776 | pronephric proximal tubule development(GO:0035776) |
| 0.3 | 1.3 | GO:0033280 | response to vitamin D(GO:0033280) vitamin D metabolic process(GO:0042359) |
| 0.3 | 1.3 | GO:0071387 | response to cortisol(GO:0051414) cellular response to cortisol stimulus(GO:0071387) |
| 0.2 | 2.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.2 | 0.8 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.2 | 2.8 | GO:0060324 | face development(GO:0060324) |
| 0.2 | 0.8 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.1 | 0.8 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.1 | 1.7 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.1 | 0.7 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.1 | 0.6 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 0.3 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.1 | 0.4 | GO:0046443 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.1 | 1.7 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.4 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.1 | 0.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.7 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 | 0.2 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.8 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.9 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 0.7 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 0.5 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.1 | 0.4 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.9 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 4.4 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.8 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.0 | 0.8 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 1.4 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.5 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.1 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 1.7 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.5 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.5 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.6 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.2 | GO:0051963 | regulation of synapse assembly(GO:0051963) positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.3 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.6 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 0.7 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.4 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.8 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 1.1 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 0.5 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.2 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.7 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 0.7 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 0.3 | GO:0016925 | protein sumoylation(GO:0016925) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 2.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.4 | GO:0031431 | Dbf4-dependent protein kinase complex(GO:0031431) |
| 0.1 | 2.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.8 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 2.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 3.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.7 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.2 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 1.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.9 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.5 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.5 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.7 | 2.2 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.6 | 2.9 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.3 | 1.3 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.3 | 0.9 | GO:0098639 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.3 | 0.8 | GO:0004953 | icosanoid receptor activity(GO:0004953) |
| 0.2 | 0.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.4 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.1 | 1.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.4 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.1 | 1.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.7 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.4 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 0.3 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 0.3 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.1 | 1.4 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 0.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.2 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 5.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 0.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 2.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.5 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.4 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 3.3 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.9 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 1.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.7 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.0 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.0 | 0.0 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 2.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 2.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.2 | 1.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 0.5 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |