Project
DANIO-CODE
Navigation
Downloads
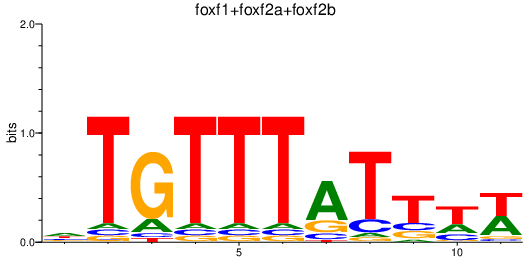
Results for foxf1+foxf2a+foxf2b
Z-value: 0.67
Transcription factors associated with foxf1+foxf2a+foxf2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxf1
|
ENSDARG00000015399 | forkhead box F1 |
|
foxf2a
|
ENSDARG00000017195 | forkhead box F2a |
|
foxf2b
|
ENSDARG00000070389 | forkhead box F2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxf2a | dr10_dc_chr2_-_844040_844078 | 0.79 | 2.8e-04 | Click! |
| foxf1 | dr10_dc_chr18_+_30868972_30869016 | 0.77 | 4.9e-04 | Click! |
Activity profile of foxf1+foxf2a+foxf2b motif
Sorted Z-values of foxf1+foxf2a+foxf2b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of foxf1+foxf2a+foxf2b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_51392394 | 1.79 |
ENSDART00000143276
|
setbp1
|
SET binding protein 1 |
| chr8_-_9081109 | 1.52 |
ENSDART00000176850
|
slc6a8
|
solute carrier family 6 (neurotransmitter transporter), member 8 |
| chr9_-_39195468 | 1.44 |
ENSDART00000014207
|
myl1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr17_+_23278879 | 1.36 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr1_+_50395721 | 1.29 |
ENSDART00000134065
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr23_-_27226280 | 1.27 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr5_-_11723101 | 1.25 |
ENSDART00000161706
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr4_-_17735989 | 1.19 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr9_+_54185607 | 1.18 |
ENSDART00000165991
|
lect1
|
leukocyte cell derived chemotaxin 1 |
| chr7_-_50827308 | 1.14 |
ENSDART00000121574
|
col4a6
|
collagen, type IV, alpha 6 |
| chr5_+_15880920 | 1.13 |
ENSDART00000168643
|
CABZ01088700.1
|
ENSDARG00000101452 |
| chr25_+_30699938 | 1.08 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr22_+_37599150 | 1.06 |
ENSDART00000065239
|
dnajc19
|
DnaJ (Hsp40) homolog, subfamily C, member 19 |
| chr7_-_22685672 | 1.06 |
ENSDART00000101447
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr1_+_41148042 | 1.05 |
ENSDART00000145170
ENSDART00000136879 |
smox
|
spermine oxidase |
| chr25_-_30845998 | 1.02 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr24_+_38371710 | 1.02 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr7_+_58443471 | 0.99 |
ENSDART00000148851
|
rps20
|
ribosomal protein S20 |
| chr15_+_28753020 | 0.98 |
ENSDART00000155815
|
nova2
|
neuro-oncological ventral antigen 2 |
| chr14_-_7774262 | 0.97 |
ENSDART00000045109
|
zgc:92242
|
zgc:92242 |
| chr5_+_45822196 | 0.97 |
|
|
|
| chr6_-_32718634 | 0.97 |
ENSDART00000175666
|
mafaa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Aa |
| chr16_+_5466342 | 0.95 |
ENSDART00000160008
|
plecb
|
plectin b |
| chr23_-_27226387 | 0.93 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr5_+_49093250 | 0.88 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr13_-_30515486 | 0.88 |
ENSDART00000139607
|
zcchc24
|
zinc finger, CCHC domain containing 24 |
| chr20_+_54289892 | 0.88 |
ENSDART00000060444
|
rps29
|
ribosomal protein S29 |
| chr19_-_29205158 | 0.86 |
ENSDART00000026992
|
sox4a
|
SRY (sex determining region Y)-box 4a |
| chr7_-_32752138 | 0.85 |
ENSDART00000052383
|
cd81a
|
CD81 molecule a |
| chr4_-_24152926 | 0.85 |
ENSDART00000123199
|
usp6nl
|
USP6 N-terminal like |
| chr14_-_16023198 | 0.85 |
|
|
|
| chr5_+_49093134 | 0.82 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr9_-_29056763 | 0.82 |
ENSDART00000060321
|
ENSDARG00000056704
|
ENSDARG00000056704 |
| chr11_-_29316162 | 0.81 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr11_+_7518953 | 0.80 |
ENSDART00000171813
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr16_+_14697969 | 0.79 |
ENSDART00000133566
|
deptor
|
DEP domain containing MTOR-interacting protein |
| chr12_+_36796886 | 0.78 |
ENSDART00000125900
|
hs3st3b1b
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1b |
| chr17_+_8166167 | 0.77 |
ENSDART00000169900
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr7_+_21521123 | 0.76 |
ENSDART00000112531
ENSDART00000100936 |
tmem88b
|
transmembrane protein 88 b |
| chr1_-_44009572 | 0.76 |
ENSDART00000144900
|
ENSDARG00000079632
|
ENSDARG00000079632 |
| chr6_-_14821305 | 0.75 |
|
|
|
| chr23_-_10202347 | 0.75 |
ENSDART00000142442
|
plxnb1a
|
plexin b1a |
| chr6_-_8501230 | 0.74 |
ENSDART00000143956
|
cavin2b
|
caveolae associated protein 2b |
| chr20_+_52740555 | 0.74 |
ENSDART00000110777
|
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr6_+_54231519 | 0.72 |
ENSDART00000149542
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr5_-_13872815 | 0.71 |
ENSDART00000164698
|
CT009570.1
|
ENSDARG00000104490 |
| chr21_+_17731439 | 0.71 |
ENSDART00000124173
|
rxraa
|
retinoid X receptor, alpha a |
| chr1_-_8430894 | 0.70 |
ENSDART00000125596
|
si:ch211-14k19.8
|
si:ch211-14k19.8 |
| chr24_+_18154733 | 0.70 |
ENSDART00000140994
|
tpk1
|
thiamin pyrophosphokinase 1 |
| chr12_+_30119269 | 0.69 |
ENSDART00000102081
|
afap1l2
|
actin filament associated protein 1-like 2 |
| chr23_-_31446156 | 0.68 |
ENSDART00000053367
|
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr5_+_31745031 | 0.66 |
ENSDART00000147132
|
c9
|
complement component 9 |
| chr17_+_24596687 | 0.66 |
ENSDART00000064739
|
rpl13a
|
ribosomal protein L13a |
| chr23_-_20332076 | 0.65 |
ENSDART00000147326
|
lamb2
|
laminin, beta 2 (laminin S) |
| chr21_+_26035166 | 0.65 |
ENSDART00000134939
|
rpl23a
|
ribosomal protein L23a |
| chr1_-_31123999 | 0.65 |
ENSDART00000152493
|
nt5c2b
|
5'-nucleotidase, cytosolic IIb |
| chr5_-_13872895 | 0.65 |
ENSDART00000164698
|
CT009570.1
|
ENSDARG00000104490 |
| chr8_-_42617646 | 0.63 |
ENSDART00000075550
|
kazald3
|
Kazal-type serine peptidase inhibitor domain 3 |
| chr5_-_43121949 | 0.63 |
ENSDART00000157093
|
si:dkey-40c11.1
|
si:dkey-40c11.1 |
| chr22_-_13139894 | 0.63 |
ENSDART00000105762
|
ahr2
|
aryl hydrocarbon receptor 2 |
| chr1_-_29706856 | 0.63 |
ENSDART00000174868
|
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr8_-_27839244 | 0.62 |
ENSDART00000136562
|
cttnbp2nlb
|
CTTNBP2 N-terminal like b |
| chr14_-_25879853 | 0.62 |
ENSDART00000014149
|
fat2
|
FAT atypical cadherin 2 |
| chr25_-_34677352 | 0.61 |
ENSDART00000115210
|
ano9a
|
anoctamin 9a |
| chr2_-_24275851 | 0.61 |
ENSDART00000121885
|
tgfbr1a
|
transforming growth factor, beta receptor 1 a |
| chr21_-_25258473 | 0.61 |
ENSDART00000101153
ENSDART00000147860 ENSDART00000087910 |
st14b
|
suppression of tumorigenicity 14 (colon carcinoma) b |
| chr2_+_5651428 | 0.60 |
ENSDART00000111220
|
mb21d2
|
Mab-21 domain containing 2 |
| chr23_+_14834670 | 0.60 |
ENSDART00000143675
|
si:rp71-79p20.2
|
si:rp71-79p20.2 |
| chr23_+_25428372 | 0.60 |
ENSDART00000147440
|
fmnl3
|
formin-like 3 |
| chr23_+_25378921 | 0.59 |
ENSDART00000143291
|
RPL41
|
ribosomal protein L41 |
| chr10_+_4987494 | 0.59 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr21_-_40694583 | 0.59 |
ENSDART00000045124
|
pomp
|
proteasome maturation protein |
| chr3_+_56008829 | 0.58 |
|
|
|
| chr11_-_21422588 | 0.58 |
|
|
|
| chr20_+_19613133 | 0.58 |
ENSDART00000152548
ENSDART00000063696 |
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr6_+_12733209 | 0.57 |
ENSDART00000167021
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr6_+_24717985 | 0.57 |
ENSDART00000165609
|
barhl2
|
BarH-like homeobox 2 |
| chr4_-_6800721 | 0.57 |
ENSDART00000099467
|
ifrd1
|
interferon-related developmental regulator 1 |
| chr10_-_42111408 | 0.57 |
ENSDART00000033121
|
morn3
|
MORN repeat containing 3 |
| chr4_+_13811932 | 0.56 |
ENSDART00000067168
|
pdzrn4
|
PDZ domain containing ring finger 4 |
| chr21_-_5852663 | 0.56 |
ENSDART00000130521
|
CACFD1
|
calcium channel flower domain containing 1 |
| chr13_+_22545318 | 0.55 |
ENSDART00000143312
|
zgc:193505
|
zgc:193505 |
| chr12_+_3875774 | 0.55 |
ENSDART00000013465
|
tbx6
|
T-box 6 |
| chr12_-_41124276 | 0.54 |
ENSDART00000172022
ENSDART00000158605 ENSDART00000162967 |
dpysl4
|
dihydropyrimidinase-like 4 |
| chr16_-_31670211 | 0.54 |
ENSDART00000138216
|
CR855311.1
|
ENSDARG00000090352 |
| chr23_+_5632124 | 0.54 |
ENSDART00000059307
|
smpd2a
|
sphingomyelin phosphodiesterase 2a, neutral membrane (neutral sphingomyelinase) |
| chr23_-_32378111 | 0.53 |
ENSDART00000143772
|
dgkaa
|
diacylglycerol kinase, alpha a |
| chr14_+_38375351 | 0.53 |
ENSDART00000175744
|
BX088707.3
|
ENSDARG00000107351 |
| chr14_+_33382973 | 0.52 |
ENSDART00000132488
|
apln
|
apelin |
| chr14_+_4169846 | 0.52 |
ENSDART00000038301
|
gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr18_+_618393 | 0.52 |
ENSDART00000159464
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr6_+_29420644 | 0.52 |
ENSDART00000065293
|
usp13
|
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr23_-_38228305 | 0.51 |
ENSDART00000131791
|
pfdn4
|
prefoldin subunit 4 |
| chr1_-_10423289 | 0.51 |
|
|
|
| chr20_+_14979154 | 0.51 |
ENSDART00000118157
|
dre-mir-199-1
|
dre-mir-199-1 |
| chr23_-_27721250 | 0.50 |
ENSDART00000059974
|
wnt10b
|
wingless-type MMTV integration site family, member 10b |
| chr5_+_18740715 | 0.50 |
ENSDART00000089173
|
atp8b5a
|
ATPase, class I, type 8B, member 5a |
| chr25_+_2909562 | 0.50 |
ENSDART00000149360
|
mpi
|
mannose phosphate isomerase |
| chr22_+_11726312 | 0.50 |
ENSDART00000155366
|
krt96
|
keratin 96 |
| chr25_-_36713728 | 0.50 |
ENSDART00000103222
|
nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr11_-_36088889 | 0.50 |
ENSDART00000146495
|
psma5
|
proteasome subunit alpha 5 |
| chr13_+_30190977 | 0.49 |
|
|
|
| chr18_+_14369543 | 0.49 |
|
|
|
| chr16_-_30606364 | 0.49 |
ENSDART00000147986
|
lmna
|
lamin A |
| chr3_-_42649955 | 0.48 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr12_+_42244935 | 0.48 |
ENSDART00000166484
|
si:ch211-221j21.3
|
si:ch211-221j21.3 |
| chr4_+_119964 | 0.48 |
ENSDART00000177177
ENSDART00000159996 |
tbk1
|
TANK-binding kinase 1 |
| chr16_-_19154920 | 0.48 |
ENSDART00000088818
|
fhod3b
|
formin homology 2 domain containing 3b |
| chr6_+_20044580 | 0.48 |
ENSDART00000129202
|
ENSDARG00000010873
|
ENSDARG00000010873 |
| chr22_+_24362712 | 0.48 |
ENSDART00000157861
|
p3h2
|
prolyl 3-hydroxylase 2 |
| chr18_+_38793580 | 0.47 |
ENSDART00000129750
|
fam214a
|
family with sequence similarity 214, member A |
| chr7_-_22685631 | 0.47 |
ENSDART00000122113
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr5_-_58162552 | 0.47 |
ENSDART00000161230
|
arhgef12b
|
Rho guanine nucleotide exchange factor (GEF) 12b |
| chr3_-_39554155 | 0.46 |
ENSDART00000015393
|
b9d1
|
B9 protein domain 1 |
| KN149955v1_+_4206 | 0.46 |
ENSDART00000167370
|
cdkn1d
|
cyclin-dependent kinase inhibitor 1D |
| chr7_+_9726412 | 0.45 |
ENSDART00000173155
|
adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr20_+_4567653 | 0.45 |
|
|
|
| chr7_+_27562912 | 0.45 |
ENSDART00000052656
|
rras2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr23_+_17294623 | 0.45 |
ENSDART00000054761
|
nol4lb
|
nucleolar protein 4-like b |
| chr25_-_13580057 | 0.45 |
ENSDART00000090226
|
znf319b
|
zinc finger protein 319b |
| chr12_+_19240407 | 0.44 |
ENSDART00000041711
|
gspt1
|
G1 to S phase transition 1 |
| chr1_-_44199728 | 0.44 |
ENSDART00000145354
|
tcirg1a
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3a |
| KN149764v1_-_1960 | 0.44 |
|
|
|
| chr4_-_25282256 | 0.44 |
ENSDART00000066936
|
tmem110l
|
transmembrane protein 110, like |
| chr6_-_57725991 | 0.43 |
ENSDART00000168708
|
cbfa2t2
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr4_+_70917419 | 0.43 |
ENSDART00000174035
|
si:cabz01054394.5
|
si:cabz01054394.5 |
| chr5_-_25509259 | 0.42 |
ENSDART00000137376
|
BX510923.1
|
ENSDARG00000094488 |
| chr16_-_12621624 | 0.42 |
ENSDART00000155829
|
cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chr4_-_6365088 | 0.42 |
ENSDART00000140100
|
MDFIC
|
MyoD family inhibitor domain containing |
| chr25_+_28717079 | 0.42 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase, muscle, b |
| chr8_+_20456215 | 0.42 |
ENSDART00000036630
|
zgc:101100
|
zgc:101100 |
| chr25_+_13607097 | 0.41 |
|
|
|
| chr6_+_45917081 | 0.41 |
ENSDART00000149450
ENSDART00000149642 |
h6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr11_+_30416953 | 0.41 |
|
|
|
| chr16_+_22672315 | 0.41 |
ENSDART00000142241
|
she
|
Src homology 2 domain containing E |
| chr23_+_13777809 | 0.41 |
ENSDART00000144386
|
uckl1b
|
uridine-cytidine kinase 1-like 1b |
| chr6_-_30696679 | 0.40 |
ENSDART00000065212
|
ttc4
|
tetratricopeptide repeat domain 4 |
| chr9_-_33330725 | 0.40 |
|
|
|
| chr4_-_11582061 | 0.40 |
ENSDART00000049066
|
net1
|
neuroepithelial cell transforming 1 |
| chr10_-_36690119 | 0.40 |
ENSDART00000077161
|
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 |
| chr5_+_34022151 | 0.40 |
ENSDART00000141338
|
enc1
|
ectodermal-neural cortex 1 |
| chr2_-_43347158 | 0.40 |
ENSDART00000141087
|
crema
|
cAMP responsive element modulator a |
| chr15_+_23615727 | 0.40 |
ENSDART00000152720
|
MARK4
|
microtubule affinity regulating kinase 4 |
| chr8_-_16630507 | 0.40 |
|
|
|
| KN149955v1_+_4134 | 0.40 |
ENSDART00000167370
|
cdkn1d
|
cyclin-dependent kinase inhibitor 1D |
| chr21_-_20728623 | 0.39 |
ENSDART00000135940
ENSDART00000002231 |
ghrb
|
growth hormone receptor b |
| chr9_-_30073641 | 0.39 |
|
|
|
| chr16_-_33637966 | 0.39 |
ENSDART00000142965
|
BX511161.1
|
ENSDARG00000092272 |
| chr17_-_32790491 | 0.39 |
ENSDART00000176858
|
cenpf
|
centromere protein F |
| chr18_-_48375325 | 0.39 |
|
|
|
| chr5_-_25509188 | 0.39 |
ENSDART00000137376
|
BX510923.1
|
ENSDARG00000094488 |
| chr18_+_29178482 | 0.38 |
ENSDART00000137587
ENSDART00000144423 |
ppfibp2a
|
PTPRF interacting protein, binding protein 2a (liprin beta 2) |
| chr20_-_29572001 | 0.38 |
ENSDART00000152906
|
scg5
|
secretogranin V |
| chr22_+_31866783 | 0.38 |
ENSDART00000159825
|
dock3
|
dedicator of cytokinesis 3 |
| chr14_+_7626822 | 0.37 |
ENSDART00000109941
|
cxxc5b
|
CXXC finger protein 5b |
| chr23_-_43595189 | 0.37 |
|
|
|
| chr10_-_43289221 | 0.37 |
ENSDART00000148293
|
tmem167a
|
transmembrane protein 167A |
| chr12_+_26376471 | 0.37 |
ENSDART00000022495
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr1_+_13779755 | 0.36 |
|
|
|
| chr13_+_38176374 | 0.36 |
ENSDART00000145777
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr21_-_9313788 | 0.36 |
|
|
|
| chr7_+_60874319 | 0.36 |
ENSDART00000110671
|
zgc:194930
|
zgc:194930 |
| chr19_-_44495755 | 0.36 |
ENSDART00000132800
|
rad21b
|
RAD21 cohesin complex component b |
| chr24_-_21324086 | 0.36 |
|
|
|
| chr12_-_9256949 | 0.36 |
ENSDART00000003805
|
pth1rb
|
parathyroid hormone 1 receptor b |
| chr7_+_19857885 | 0.35 |
ENSDART00000146335
|
zgc:114045
|
zgc:114045 |
| chr21_-_30144979 | 0.35 |
ENSDART00000065447
|
hnrnph1l
|
heterogeneous nuclear ribonucleoprotein H1, like |
| chr8_+_49582361 | 0.35 |
ENSDART00000108613
|
rasef
|
RAS and EF-hand domain containing |
| chr16_-_26083913 | 0.35 |
ENSDART00000166681
|
frrs1l
|
ferric-chelate reductase 1-like |
| chr25_-_3766929 | 0.34 |
ENSDART00000043172
|
tmem258
|
transmembrane protein 258 |
| chr12_+_27612667 | 0.34 |
ENSDART00000153256
|
cacna1g
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr15_-_20997836 | 0.34 |
ENSDART00000152648
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr6_-_40660116 | 0.34 |
ENSDART00000154359
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr5_+_62987426 | 0.34 |
ENSDART00000178937
|
dnm1b
|
dynamin 1b |
| chr12_+_17744208 | 0.34 |
|
|
|
| chr20_-_47932043 | 0.34 |
ENSDART00000085624
|
ipo13
|
importin 13 |
| chr14_-_6939857 | 0.34 |
ENSDART00000108796
|
stox2b
|
storkhead box 2b |
| chr7_+_19857748 | 0.33 |
ENSDART00000146335
|
zgc:114045
|
zgc:114045 |
| chr17_+_8166308 | 0.33 |
ENSDART00000172443
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr14_+_34626233 | 0.33 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr5_+_38903966 | 0.33 |
ENSDART00000121460
|
prdm8b
|
PR domain containing 8b |
| chr24_-_38496534 | 0.33 |
ENSDART00000140739
|
lrrc4bb
|
leucine rich repeat containing 4Bb |
| chr1_+_44009649 | 0.33 |
ENSDART00000131296
|
ssrp1b
|
structure specific recognition protein 1b |
| chr2_+_39653315 | 0.33 |
ENSDART00000077108
|
zgc:136870
|
zgc:136870 |
| chr21_-_26035057 | 0.33 |
ENSDART00000141382
|
rab34b
|
RAB34, member RAS oncogene family b |
| chr11_-_1792616 | 0.33 |
|
|
|
| chr8_+_5174495 | 0.33 |
ENSDART00000155816
|
ppp2r2aa
|
protein phosphatase 2, regulatory subunit B, alpha a |
| chr4_-_1975473 | 0.33 |
ENSDART00000111858
|
mrpl42
|
mitochondrial ribosomal protein L42 |
| chr16_-_28921444 | 0.33 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr9_+_6684835 | 0.32 |
|
|
|
| chr24_+_30343717 | 0.32 |
ENSDART00000162377
|
FP085414.1
|
ENSDARG00000100270 |
| chr18_-_43890514 | 0.32 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr15_-_7327976 | 0.32 |
|
|
|
| chr23_+_35405877 | 0.32 |
ENSDART00000159218
|
acp1
|
acid phosphatase 1, soluble |
| chr21_+_30512356 | 0.32 |
ENSDART00000109719
|
rab38c
|
RAB38c, member of RAS oncogene family |
| chr4_-_5589261 | 0.31 |
ENSDART00000017349
|
vegfab
|
vascular endothelial growth factor Ab |
| chr5_+_51992974 | 0.31 |
ENSDART00000170341
|
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr5_-_43122200 | 0.31 |
ENSDART00000157093
|
si:dkey-40c11.1
|
si:dkey-40c11.1 |
| chr11_-_16080475 | 0.31 |
ENSDART00000027014
|
rab7
|
RAB7, member RAS oncogene family |
| chr25_+_32348377 | 0.31 |
ENSDART00000162188
|
ETFA
|
electron transfer flavoprotein alpha subunit |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.3 | 0.9 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine diphosphate metabolic process(GO:0042357) thiamine-containing compound metabolic process(GO:0042723) |
| 0.3 | 0.9 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.3 | 0.8 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.3 | 1.3 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.3 | 1.0 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.2 | 1.6 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.2 | 0.8 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.2 | 0.9 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.2 | 2.0 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.2 | 0.5 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.2 | 0.5 | GO:0090171 | cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) chondrocyte morphogenesis(GO:0090171) |
| 0.2 | 0.6 | GO:0042698 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.2 | 0.6 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.1 | 1.1 | GO:0032048 | cardiolipin metabolic process(GO:0032048) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.1 | 1.0 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.7 | GO:0071333 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.1 | 0.3 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 0.7 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 0.3 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) regulation of nonmotile primary cilium assembly(GO:1902855) |
| 0.1 | 0.6 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.1 | 0.7 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 1.8 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.1 | 0.4 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.3 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.5 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.1 | 0.3 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 0.8 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.2 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.1 | 0.5 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.1 | 0.3 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.7 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.5 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.1 | 0.2 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 0.2 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 0.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.5 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.1 | 0.4 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 1.0 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 0.5 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.1 | 0.3 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.2 | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.1 | 0.2 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.1 | 0.2 | GO:0032635 | interleukin-6 production(GO:0032635) regulation of interleukin-6 production(GO:0032675) |
| 0.1 | 0.3 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.3 | GO:0030317 | sperm motility(GO:0030317) |
| 0.1 | 0.4 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) CTP salvage(GO:0044211) |
| 0.0 | 0.5 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.6 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.6 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0046874 | quinolinate biosynthetic process(GO:0019805) quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) |
| 0.0 | 0.2 | GO:0070198 | protein localization to chromosome, telomeric region(GO:0070198) |
| 0.0 | 0.2 | GO:0008291 | neuromuscular synaptic transmission(GO:0007274) acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.2 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.4 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.3 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 1.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.1 | GO:0015865 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.3 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.8 | GO:0032963 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.0 | 0.3 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 1.7 | GO:0072331 | signal transduction by p53 class mediator(GO:0072331) |
| 0.0 | 0.7 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.2 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.0 | 0.9 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 1.5 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.2 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.4 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.0 | 0.2 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.0 | 0.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.3 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.3 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.3 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) catecholamine secretion(GO:0050432) regulation of catecholamine secretion(GO:0050433) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.3 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.6 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.3 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.9 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.4 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.7 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.2 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.1 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.2 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.4 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.3 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.0 | 0.2 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.3 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.4 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.1 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.3 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.8 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.2 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.2 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0098594 | mucin granule(GO:0098594) |
| 0.3 | 1.1 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.3 | 1.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.2 | 0.7 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 0.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.4 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 1.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.4 | GO:0030892 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.7 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 1.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.7 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.3 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 0.7 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 0.3 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.3 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 1.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 1.5 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.5 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 1.3 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.5 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.4 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 1.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.9 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.1 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.3 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.4 | 1.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.3 | 1.0 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.2 | 0.8 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.2 | 0.8 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 1.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.6 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 0.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.7 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.4 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.4 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.1 | 0.8 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.1 | 0.4 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.9 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.1 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.6 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 1.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.9 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.5 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.7 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.3 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 1.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.8 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.1 | 0.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.4 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 2.1 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 1.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 2.1 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 3.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.6 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 1.5 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.4 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.4 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 0.1 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.0 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.9 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 0.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 1.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 0.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 0.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 3.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.8 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.7 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |