Project
DANIO-CODE
Navigation
Downloads
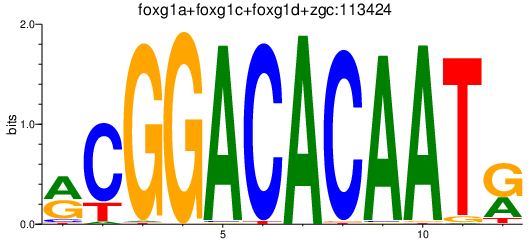
Results for foxg1a+foxg1c+foxg1d+zgc:113424
Z-value: 0.66
Transcription factors associated with foxg1a+foxg1c+foxg1d+zgc:113424
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zgc
|
ENSDARG00000035680 | 113424 |
|
foxg1c
|
ENSDARG00000068380 | forkhead box G1c |
|
foxg1d
|
ENSDARG00000070053 | forkhead box G1d |
|
foxg1a
|
ENSDARG00000070769 | forkhead box G1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxg1a | dr10_dc_chr17_-_29102320_29102403 | 0.93 | 2.7e-07 | Click! |
| zgc:113424 | dr10_dc_chr19_-_35411993_35412339 | -0.89 | 3.3e-06 | Click! |
| foxg1d | dr10_dc_chr13_+_47175_47180 | 0.82 | 9.3e-05 | Click! |
Activity profile of foxg1a+foxg1c+foxg1d+zgc:113424 motif
Sorted Z-values of foxg1a+foxg1c+foxg1d+zgc:113424 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of foxg1a+foxg1c+foxg1d+zgc:113424
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_42032702 | 2.59 |
|
|
|
| chr2_+_49358871 | 2.24 |
ENSDART00000179089
|
sema6ba
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Ba |
| chr1_-_50066633 | 2.04 |
|
|
|
| chr15_-_23587330 | 1.96 |
ENSDART00000167246
|
hmbsb
|
hydroxymethylbilane synthase, b |
| chr6_-_18891908 | 1.89 |
ENSDART00000074327
|
igfbp2a
|
insulin-like growth factor binding protein 2a |
| chr1_+_45063098 | 1.85 |
ENSDART00000084512
|
pkn1a
|
protein kinase N1a |
| chr9_+_54692834 | 1.79 |
|
|
|
| chr11_-_26428979 | 1.72 |
ENSDART00000111539
|
efcc1
|
EF-hand and coiled-coil domain containing 1 |
| chr6_+_47845002 | 1.64 |
ENSDART00000140943
|
padi2
|
peptidyl arginine deiminase, type II |
| chr23_+_36832325 | 1.36 |
|
|
|
| chr21_+_43674754 | 1.35 |
ENSDART00000136025
|
tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr24_-_2278409 | 1.32 |
|
|
|
| chr23_+_6043862 | 1.30 |
|
|
|
| chr1_+_51706536 | 1.29 |
|
|
|
| chr7_-_72500748 | 1.22 |
ENSDART00000160523
|
CU929444.1
|
ENSDARG00000099109 |
| chr18_+_16257606 | 1.20 |
ENSDART00000142584
|
alx1
|
ALX homeobox 1 |
| chr20_+_34417665 | 1.18 |
ENSDART00000153207
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr7_-_8635687 | 1.17 |
ENSDART00000081620
|
vax2
|
ventral anterior homeobox 2 |
| chr24_-_36428001 | 1.08 |
ENSDART00000048046
|
naglu
|
N-acetylglucosaminidase, alpha |
| chr21_-_3301751 | 1.00 |
ENSDART00000009740
|
smad7
|
SMAD family member 7 |
| chr6_+_52064243 | 0.95 |
ENSDART00000153468
|
abraa
|
actin binding Rho activating protein a |
| chr24_-_36427944 | 0.90 |
ENSDART00000048046
|
naglu
|
N-acetylglucosaminidase, alpha |
| chr6_-_16156494 | 0.89 |
|
|
|
| chr19_-_22956977 | 0.85 |
ENSDART00000019505
|
pleca
|
plectin a |
| chr7_+_72532825 | 0.75 |
ENSDART00000175316
|
FO818686.1
|
ENSDARG00000108531 |
| chr24_+_19447486 | 0.71 |
ENSDART00000152026
|
slco5a1a
|
solute carrier organic anion transporter family member 5A1a |
| chr25_-_35159840 | 0.71 |
ENSDART00000148718
ENSDART00000164189 ENSDART00000027174 |
lrrk2
|
leucine-rich repeat kinase 2 |
| chr5_-_37521979 | 0.70 |
ENSDART00000141791
ENSDART00000170528 |
si:ch211-284e13.6
|
si:ch211-284e13.6 |
| chr18_-_34194840 | 0.62 |
ENSDART00000015079
ENSDART00000133400 |
slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr6_+_3703684 | 0.61 |
ENSDART00000013743
|
gorasp2
|
golgi reassembly stacking protein 2 |
| chr24_+_37752791 | 0.59 |
ENSDART00000158181
|
wdr24
|
WD repeat domain 24 |
| chr1_+_51706568 | 0.59 |
|
|
|
| chr6_+_11594333 | 0.59 |
ENSDART00000109552
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr18_+_18115168 | 0.57 |
ENSDART00000145342
|
cbln1
|
cerebellin 1 precursor |
| chr19_-_22957035 | 0.49 |
ENSDART00000019505
|
pleca
|
plectin a |
| chr3_+_23118171 | 0.47 |
ENSDART00000156032
|
ppp1r9ba
|
protein phosphatase 1, regulatory subunit 9Ba |
| chr6_+_47845098 | 0.45 |
ENSDART00000127766
|
padi2
|
peptidyl arginine deiminase, type II |
| chr24_+_18141977 | 0.45 |
ENSDART00000055443
|
tpk1
|
thiamin pyrophosphokinase 1 |
| chr7_+_60992745 | 0.44 |
ENSDART00000083255
|
adam19a
|
ADAM metallopeptidase domain 19a |
| chr11_+_13000957 | 0.44 |
ENSDART00000161532
|
zfyve9b
|
zinc finger, FYVE domain containing 9b |
| chr13_+_24417183 | 0.42 |
ENSDART00000177677
|
lgalslb
|
lectin, galactoside-binding-like b |
| chr22_-_12723704 | 0.42 |
ENSDART00000080741
|
plcd4a
|
phospholipase C, delta 4a |
| chr17_+_19514507 | 0.36 |
|
|
|
| chr11_-_42263721 | 0.33 |
ENSDART00000130573
|
atp6ap1la
|
ATPase, H+ transporting, lysosomal accessory protein 1-like a |
| chr2_-_7375138 | 0.33 |
ENSDART00000146434
|
zgc:153115
|
zgc:153115 |
| chr7_+_72533052 | 0.27 |
ENSDART00000175316
|
FO818686.1
|
ENSDARG00000108531 |
| chr15_-_8786861 | 0.26 |
ENSDART00000154171
|
arhgap35a
|
Rho GTPase activating protein 35a |
| chr2_-_23331500 | 0.24 |
ENSDART00000146497
|
mllt1b
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 1b |
| chr10_-_38206238 | 0.21 |
ENSDART00000174882
|
FP016248.1
|
ENSDARG00000107804 |
| chr18_-_5656654 | 0.14 |
ENSDART00000142945
ENSDART00000063024 |
bloc1s6
|
biogenesis of lysosomal organelles complex-1, subunit 6, pallidin |
| chr3_+_41584507 | 0.13 |
ENSDART00000154401
|
chst12a
|
carbohydrate (chondroitin 4) sulfotransferase 12a |
| chr2_+_35898064 | 0.11 |
ENSDART00000004277
|
lamc1
|
laminin, gamma 1 |
| chr4_+_9359769 | 0.10 |
ENSDART00000128937
|
ipo8
|
importin 8 |
| chr10_-_38206060 | 0.08 |
ENSDART00000174882
|
FP016248.1
|
ENSDARG00000107804 |
| chr18_+_5657314 | 0.05 |
ENSDART00000007797
|
slc30a4
|
solute carrier family 30 (zinc transporter), member 4 |
| chr24_-_37752716 | 0.04 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr10_+_29964412 | 0.03 |
ENSDART00000117710
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr25_-_26230031 | 0.01 |
ENSDART00000154917
|
ciartb
|
circadian associated repressor of transcription b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.5 | 1.9 | GO:0040014 | regulation of multicellular organism growth(GO:0040014) |
| 0.4 | 1.2 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.2 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 1.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 0.7 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 1.2 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.1 | 0.4 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine diphosphate metabolic process(GO:0042357) thiamine-containing compound metabolic process(GO:0042723) |
| 0.1 | 1.2 | GO:1902254 | regulation of retinal ganglion cell axon guidance(GO:0090259) negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.1 | 0.4 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.1 | 1.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.0 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 2.2 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 1.0 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.6 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.1 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.0 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 1.9 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.5 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.0 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.3 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.2 | 0.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.2 | 1.9 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 1.2 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.1 | 1.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.6 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 1.9 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 1.0 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.4 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 2.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.3 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.7 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.4 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |