Project
DANIO-CODE
Navigation
Downloads
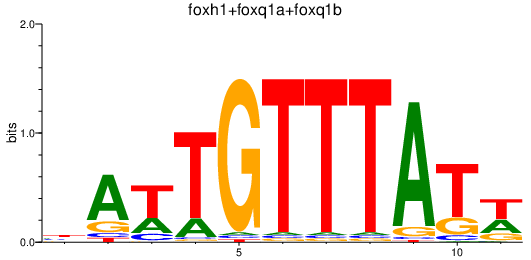
Results for foxh1+foxq1a+foxq1b
Z-value: 0.45
Transcription factors associated with foxh1+foxq1a+foxq1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxq1a
|
ENSDARG00000030896 | forkhead box Q1a |
|
foxq1b
|
ENSDARG00000055395 | forkhead box Q1b |
|
foxh1
|
ENSDARG00000055630 | forkhead box H1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxq1b | dr10_dc_chr20_+_26784812_26784820 | -0.44 | 8.9e-02 | Click! |
| foxh1 | dr10_dc_chr12_-_13690539_13690686 | 0.41 | 1.2e-01 | Click! |
Activity profile of foxh1+foxq1a+foxq1b motif
Sorted Z-values of foxh1+foxq1a+foxq1b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of foxh1+foxq1a+foxq1b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_33391554 | 0.83 |
ENSDART00000024104
|
mycn
|
MYCN proto-oncogene, bHLH transcription factor |
| chr18_+_40365208 | 0.62 |
ENSDART00000167134
|
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr14_+_34154895 | 0.49 |
ENSDART00000147756
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr9_+_31471391 | 0.48 |
ENSDART00000010774
|
zic2a
|
zic family member 2 (odd-paired homolog, Drosophila), a |
| chr18_+_40364675 | 0.47 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr7_+_29969936 | 0.43 |
ENSDART00000109243
|
sema4bb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Bb |
| chr12_-_34786844 | 0.39 |
ENSDART00000027379
|
bicral
|
BRD4 interacting chromatin remodeling complex associated protein like |
| chr9_-_54126121 | 0.38 |
ENSDART00000126314
|
pcdh8
|
protocadherin 8 |
| chr11_-_40415291 | 0.38 |
|
|
|
| chr11_-_22143133 | 0.37 |
ENSDART00000159681
|
tfeb
|
transcription factor EB |
| chr7_-_50827308 | 0.37 |
ENSDART00000121574
|
col4a6
|
collagen, type IV, alpha 6 |
| chr9_+_17298410 | 0.36 |
ENSDART00000048548
|
scel
|
sciellin |
| chr12_-_34614931 | 0.36 |
ENSDART00000152876
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr16_-_9785057 | 0.35 |
ENSDART00000113724
|
mal2
|
mal, T-cell differentiation protein 2 (gene/pseudogene) |
| chr16_-_17148579 | 0.35 |
ENSDART00000138715
|
plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr4_+_22020246 | 0.34 |
ENSDART00000112035
ENSDART00000127664 |
myf5
|
myogenic factor 5 |
| chr11_+_37345024 | 0.34 |
ENSDART00000077496
|
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr16_-_30606364 | 0.33 |
ENSDART00000147986
|
lmna
|
lamin A |
| chr24_-_24306469 | 0.30 |
ENSDART00000154149
|
BX323067.1
|
ENSDARG00000097984 |
| chr2_-_44429891 | 0.29 |
ENSDART00000163040
ENSDART00000056372 ENSDART00000109251 ENSDART00000166923 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr18_-_44323435 | 0.29 |
ENSDART00000098599
|
si:ch211-151h10.2
|
si:ch211-151h10.2 |
| chr1_-_44888989 | 0.27 |
ENSDART00000135089
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr19_-_27807312 | 0.26 |
|
|
|
| chr11_+_24919694 | 0.25 |
ENSDART00000162618
ENSDART00000128126 |
trpc4apa
|
transient receptor potential cation channel, subfamily C, member 4 associated protein a |
| chr1_+_8605984 | 0.25 |
ENSDART00000055009
|
uncx4.1
|
Unc4.1 homeobox (C. elegans) |
| chr7_-_52848084 | 0.23 |
ENSDART00000172179
ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr25_-_3521512 | 0.23 |
ENSDART00000171863
ENSDART00000166363 |
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr7_+_29970029 | 0.23 |
ENSDART00000109243
|
sema4bb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Bb |
| chr13_-_25067585 | 0.21 |
ENSDART00000159585
|
adka
|
adenosine kinase a |
| chr15_+_20907421 | 0.21 |
ENSDART00000153726
|
ulk2
|
unc-51 like autophagy activating kinase 2 |
| chr7_+_15623852 | 0.20 |
ENSDART00000161608
|
pax6b
|
paired box 6b |
| chr18_+_19659195 | 0.20 |
ENSDART00000100569
|
smad6b
|
SMAD family member 6b |
| chr10_+_14530451 | 0.20 |
ENSDART00000026383
|
sigmar1
|
sigma non-opioid intracellular receptor 1 |
| chr19_+_20183210 | 0.20 |
ENSDART00000164968
|
hoxa4a
|
homeobox A4a |
| chr22_+_14092247 | 0.18 |
ENSDART00000013575
|
bzw1a
|
basic leucine zipper and W2 domains 1a |
| chr10_+_4987494 | 0.18 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr7_+_9726412 | 0.18 |
ENSDART00000173155
|
adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr13_-_3022193 | 0.18 |
ENSDART00000050934
|
pkdcca
|
protein kinase domain containing, cytoplasmic a |
| chr1_+_35058017 | 0.18 |
ENSDART00000085051
|
hhip
|
hedgehog interacting protein |
| chr12_-_22277951 | 0.18 |
|
|
|
| chr19_+_26386711 | 0.18 |
ENSDART00000089836
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr23_+_25428372 | 0.17 |
ENSDART00000147440
|
fmnl3
|
formin-like 3 |
| chr25_+_18460166 | 0.17 |
ENSDART00000073726
|
cav2
|
caveolin 2 |
| chr11_-_9479689 | 0.17 |
ENSDART00000175829
|
CU179643.1
|
ENSDARG00000086489 |
| chr17_+_19003200 | 0.17 |
|
|
|
| chr16_+_29623958 | 0.17 |
ENSDART00000088146
|
ensab
|
endosulfine alpha b |
| chr7_-_25794648 | 0.16 |
|
|
|
| chr21_+_28465617 | 0.16 |
ENSDART00000140229
|
otub1a
|
OTU deubiquitinase, ubiquitin aldehyde binding 1a |
| chr2_+_33352680 | 0.16 |
ENSDART00000126135
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr3_-_41944723 | 0.16 |
ENSDART00000083111
|
ttyh3a
|
tweety family member 3a |
| chr11_-_16080475 | 0.15 |
ENSDART00000027014
|
rab7
|
RAB7, member RAS oncogene family |
| chr5_+_49093250 | 0.15 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr13_-_3022033 | 0.15 |
ENSDART00000050934
|
pkdcca
|
protein kinase domain containing, cytoplasmic a |
| chr4_-_6558687 | 0.15 |
ENSDART00000150774
|
foxp2
|
forkhead box P2 |
| chr7_-_18295101 | 0.15 |
ENSDART00000173969
|
rgs12a
|
regulator of G protein signaling 12a |
| chr14_-_44794961 | 0.15 |
ENSDART00000172805
|
CABZ01074913.1
|
ENSDARG00000105401 |
| chr14_+_28180174 | 0.14 |
ENSDART00000161852
|
stag2b
|
stromal antigen 2b |
| chr16_-_13927684 | 0.14 |
ENSDART00000146719
|
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr3_+_23580269 | 0.13 |
ENSDART00000151584
|
hoxb4a
|
homeobox B4a |
| chr3_+_40147509 | 0.13 |
ENSDART00000017304
|
cpsf4
|
cleavage and polyadenylation specific factor 4 |
| chr11_-_25019298 | 0.13 |
ENSDART00000130477
ENSDART00000079578 |
snai1a
|
snail family zinc finger 1a |
| chr16_-_13927947 | 0.13 |
ENSDART00000144856
|
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr22_-_29957440 | 0.13 |
ENSDART00000019786
ENSDART00000159813 |
smc3
|
structural maintenance of chromosomes 3 |
| chr11_-_9479592 | 0.13 |
ENSDART00000175829
|
CU179643.1
|
ENSDARG00000086489 |
| chr3_+_56008829 | 0.13 |
|
|
|
| chr1_+_25005968 | 0.13 |
ENSDART00000054228
|
lrata
|
lecithin retinol acyltransferase a (phosphatidylcholine-retinol O-acyltransferase) |
| chr2_+_31974269 | 0.13 |
ENSDART00000012413
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr5_-_62816208 | 0.13 |
ENSDART00000097325
|
c5
|
complement component 5 |
| chr11_-_25019165 | 0.13 |
ENSDART00000130477
ENSDART00000079578 |
snai1a
|
snail family zinc finger 1a |
| chr7_+_25794014 | 0.12 |
ENSDART00000174010
ENSDART00000173815 |
CT027989.1
|
ENSDARG00000105638 |
| chr18_+_5899355 | 0.12 |
|
|
|
| chr16_-_42235969 | 0.12 |
ENSDART00000076030
|
fbl
|
fibrillarin |
| chr5_+_49093134 | 0.12 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| KN150001v1_+_15017 | 0.12 |
|
|
|
| chr25_+_3521763 | 0.11 |
ENSDART00000160017
|
CABZ01058261.1
|
ENSDARG00000099194 |
| chr19_+_46647893 | 0.11 |
ENSDART00000162785
ENSDART00000171251 ENSDART00000164938 |
mapk15
|
mitogen-activated protein kinase 15 |
| chr25_-_19927119 | 0.11 |
ENSDART00000174776
|
cnot4a
|
CCR4-NOT transcription complex, subunit 4a |
| chr3_-_44992432 | 0.11 |
ENSDART00000167173
|
gipc1
|
GIPC PDZ domain containing family, member 1 |
| chr19_+_42492029 | 0.11 |
ENSDART00000150949
|
nfyc
|
nuclear transcription factor Y, gamma |
| chr12_-_27496942 | 0.10 |
ENSDART00000066282
|
dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr2_-_17564676 | 0.10 |
ENSDART00000144251
|
artnb
|
artemin b |
| chr22_-_20963723 | 0.10 |
|
|
|
| chr10_-_20630768 | 0.10 |
|
|
|
| chr10_-_14530340 | 0.10 |
ENSDART00000101298
ENSDART00000138161 |
galt
|
galactose-1-phosphate uridylyltransferase |
| chr14_+_34626233 | 0.09 |
ENSDART00000004550
|
rnf145a
|
ring finger protein 145a |
| chr11_-_11689402 | 0.09 |
|
|
|
| chr6_+_33896684 | 0.09 |
ENSDART00000165710
|
gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr17_-_18869111 | 0.09 |
ENSDART00000080029
|
ak7b
|
adenylate kinase 7b |
| chr23_-_6442213 | 0.09 |
ENSDART00000112667
|
ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr22_+_39067994 | 0.09 |
|
|
|
| chr17_+_38314814 | 0.09 |
ENSDART00000017493
|
nkx2.1
|
NK2 homeobox 1 |
| chr3_+_59573755 | 0.08 |
ENSDART00000102014
|
fam195b
|
family with sequence similarity 195, member B |
| chr5_-_25509310 | 0.08 |
ENSDART00000137376
|
BX510923.1
|
ENSDARG00000094488 |
| chr15_+_23615727 | 0.08 |
ENSDART00000152720
|
MARK4
|
microtubule affinity regulating kinase 4 |
| chr11_+_7570326 | 0.08 |
ENSDART00000091550
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr12_-_34615030 | 0.08 |
ENSDART00000152876
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr8_+_25015374 | 0.08 |
ENSDART00000140617
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr3_+_17913109 | 0.08 |
ENSDART00000112384
|
ankrd40
|
ankyrin repeat domain 40 |
| chr1_+_23385409 | 0.08 |
ENSDART00000014608
|
mab21l2
|
mab-21-like 2 |
| chr24_+_26183653 | 0.07 |
ENSDART00000003884
|
mynn
|
myoneurin |
| chr13_+_18376642 | 0.07 |
ENSDART00000142622
|
si:ch211-198a12.6
|
si:ch211-198a12.6 |
| chr17_+_32670368 | 0.07 |
ENSDART00000142449
|
ctsba
|
cathepsin Ba |
| chr16_+_29558320 | 0.07 |
ENSDART00000011497
|
ctsk
|
cathepsin K |
| chr11_+_24919823 | 0.07 |
ENSDART00000162618
ENSDART00000128126 |
trpc4apa
|
transient receptor potential cation channel, subfamily C, member 4 associated protein a |
| chr4_+_119964 | 0.07 |
ENSDART00000177177
ENSDART00000159996 |
tbk1
|
TANK-binding kinase 1 |
| chr18_+_14725148 | 0.07 |
ENSDART00000146128
|
uri1
|
URI1, prefoldin-like chaperone |
| chr10_+_15398004 | 0.07 |
|
|
|
| chr18_+_30443201 | 0.07 |
ENSDART00000140908
|
gse1
|
Gse1 coiled-coil protein |
| chr7_-_20565207 | 0.07 |
ENSDART00000052977
|
cldn15a
|
claudin 15a |
| chr14_-_24113237 | 0.06 |
ENSDART00000125923
|
cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr5_-_3562233 | 0.06 |
ENSDART00000143250
|
si:ch1073-189o9.1
|
si:ch1073-189o9.1 |
| chr10_+_572931 | 0.06 |
ENSDART00000129856
ENSDART00000110384 |
smad4a
|
SMAD family member 4a |
| chr4_+_15012460 | 0.06 |
ENSDART00000124452
|
ube2h
|
ubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast) |
| chr19_-_31378033 | 0.06 |
|
|
|
| chr7_+_15623685 | 0.06 |
ENSDART00000146704
|
pax6b
|
paired box 6b |
| chr25_+_24193604 | 0.06 |
ENSDART00000083407
|
b4galnt4a
|
beta-1,4-N-acetyl-galactosaminyl transferase 4a |
| chr11_+_37345219 | 0.06 |
ENSDART00000172899
|
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr5_-_25509259 | 0.06 |
ENSDART00000137376
|
BX510923.1
|
ENSDARG00000094488 |
| chr18_+_20044862 | 0.06 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr18_+_20045221 | 0.06 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr18_-_9980662 | 0.05 |
|
|
|
| chr25_-_7801166 | 0.05 |
ENSDART00000014274
|
glcea
|
glucuronic acid epimerase a |
| chr23_-_27930478 | 0.05 |
ENSDART00000043462
|
acvrl1
|
activin A receptor type II-like 1 |
| chr18_+_14725283 | 0.05 |
ENSDART00000146128
|
uri1
|
URI1, prefoldin-like chaperone |
| chr19_+_7072033 | 0.05 |
ENSDART00000158758
ENSDART00000160482 |
kifc1
|
kinesin family member C1 |
| KN150001v1_+_14939 | 0.05 |
|
|
|
| chr3_+_23580216 | 0.05 |
ENSDART00000151584
|
hoxb4a
|
homeobox B4a |
| chr4_-_6558919 | 0.05 |
ENSDART00000142087
|
foxp2
|
forkhead box P2 |
| chr12_-_34614903 | 0.05 |
ENSDART00000152876
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| KN149681v1_-_46883 | 0.05 |
|
|
|
| chr23_+_36019989 | 0.05 |
ENSDART00000103134
ENSDART00000139319 |
hoxc5a
|
homeobox C5a |
| chr8_-_11286437 | 0.04 |
ENSDART00000008215
|
pip5k1bb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta b |
| chr3_-_49104540 | 0.04 |
ENSDART00000138503
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr7_+_9045376 | 0.04 |
ENSDART00000128530
|
snrpa1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr22_-_27252706 | 0.04 |
ENSDART00000019442
|
xpnpep1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr18_+_13108810 | 0.04 |
ENSDART00000161153
|
GAN
|
gigaxonin |
| chr7_-_25805806 | 0.04 |
ENSDART00000101120
|
zgc:92664
|
zgc:92664 |
| chr21_-_20728623 | 0.04 |
ENSDART00000135940
ENSDART00000002231 |
ghrb
|
growth hormone receptor b |
| chr3_-_42649907 | 0.04 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr17_-_42778156 | 0.04 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr10_+_4987461 | 0.04 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr9_+_3416651 | 0.04 |
ENSDART00000019910
|
dlx1a
|
distal-less homeobox 1a |
| chr12_+_19286427 | 0.04 |
ENSDART00000114248
|
snx29
|
sorting nexin 29 |
| chr23_-_38228305 | 0.04 |
ENSDART00000131791
|
pfdn4
|
prefoldin subunit 4 |
| chr9_-_30073641 | 0.04 |
|
|
|
| chr16_+_35582277 | 0.04 |
ENSDART00000167001
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr23_+_36019758 | 0.04 |
ENSDART00000103134
ENSDART00000139319 |
hoxc5a
|
homeobox C5a |
| chr5_+_37380972 | 0.04 |
|
|
|
| chr3_+_34104493 | 0.03 |
ENSDART00000124631
|
znf207a
|
zinc finger protein 207, a |
| chr2_-_27892824 | 0.03 |
|
|
|
| chr5_-_13266357 | 0.03 |
ENSDART00000134064
|
mxd1
|
MAX dimerization protein 1 |
| chr22_-_29957382 | 0.03 |
ENSDART00000019786
ENSDART00000159813 |
smc3
|
structural maintenance of chromosomes 3 |
| chr18_-_34180716 | 0.03 |
ENSDART00000059400
|
c18h3orf33
|
c18h3orf33 homolog (H. sapiens) |
| chr7_+_51883949 | 0.03 |
ENSDART00000130282
|
katnb1
|
katanin p80 (WD repeat containing) subunit B 1 |
| chr9_-_21257082 | 0.03 |
ENSDART00000124533
|
tbx15
|
T-box 15 |
| chr10_+_34372205 | 0.03 |
ENSDART00000110121
|
stard13a
|
StAR-related lipid transfer (START) domain containing 13a |
| chr7_-_60046429 | 0.03 |
ENSDART00000127518
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr21_+_26696853 | 0.03 |
ENSDART00000168379
|
pcxa
|
pyruvate carboxylase a |
| chr10_+_573116 | 0.03 |
ENSDART00000129856
ENSDART00000110384 |
smad4a
|
SMAD family member 4a |
| chr15_+_29729531 | 0.02 |
ENSDART00000135540
|
nrip1a
|
nuclear receptor interacting protein 1a |
| chr3_-_39554155 | 0.02 |
ENSDART00000015393
|
b9d1
|
B9 protein domain 1 |
| chr13_-_23838192 | 0.02 |
|
|
|
| chr12_+_22701981 | 0.02 |
ENSDART00000153199
|
afap1
|
actin filament associated protein 1 |
| chr23_+_2256243 | 0.02 |
ENSDART00000126768
|
cc2d2a
|
coiled-coil and C2 domain containing 2A |
| chr6_+_27524211 | 0.02 |
ENSDART00000079397
|
ryk
|
receptor-like tyrosine kinase |
| chr8_+_10824790 | 0.02 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr23_-_3813451 | 0.02 |
|
|
|
| chr19_-_22184035 | 0.02 |
|
|
|
| chr20_-_20455731 | 0.02 |
ENSDART00000170488
ENSDART00000047580 |
hif1ab
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) b |
| chr5_-_28167409 | 0.02 |
ENSDART00000158299
|
traf2a
|
Tnf receptor-associated factor 2a |
| chr4_-_9851224 | 0.02 |
ENSDART00000080702
|
glt8d2
|
glycosyltransferase 8 domain containing 2 |
| chr21_+_21337628 | 0.02 |
ENSDART00000136325
|
rtn2b
|
reticulon 2b |
| chr7_+_13386796 | 0.01 |
ENSDART00000157870
|
clpxa
|
caseinolytic mitochondrial matrix peptidase chaperone subunit a |
| chr15_+_16249091 | 0.01 |
ENSDART00000147900
|
rnmtl1a
|
RNA methyltransferase like 1a |
| chr1_+_52686770 | 0.01 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr14_-_44794907 | 0.01 |
ENSDART00000172805
|
CABZ01074913.1
|
ENSDARG00000105401 |
| chr4_-_6558869 | 0.01 |
ENSDART00000142087
|
foxp2
|
forkhead box P2 |
| chr20_-_16256762 | 0.01 |
ENSDART00000021550
|
ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr19_-_48732039 | 0.01 |
ENSDART00000169577
|
si:ch73-359m17.2
|
si:ch73-359m17.2 |
| chr1_+_54005969 | 0.01 |
ENSDART00000077578
|
knop1
|
lysine-rich nucleolar protein 1 |
| chr16_-_42557446 | 0.01 |
ENSDART00000109259
|
smarcc1a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1a |
| chr24_-_23178218 | 0.01 |
|
|
|
| chr7_-_60046465 | 0.01 |
ENSDART00000127518
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr8_+_25015325 | 0.01 |
ENSDART00000140617
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr22_-_22139099 | 0.01 |
|
|
|
| chr22_+_39067567 | 0.01 |
|
|
|
| chr17_+_15527525 | 0.01 |
ENSDART00000159426
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr14_-_1080574 | 0.01 |
ENSDART00000169090
ENSDART00000158905 ENSDART00000159907 |
ENSDARG00000098694
|
ENSDARG00000098694 |
| chr2_+_30439889 | 0.00 |
ENSDART00000139871
|
socs6b
|
suppressor of cytokine signaling 6b |
| chr1_+_52686829 | 0.00 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr7_-_60046624 | 0.00 |
ENSDART00000127518
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr14_-_24113324 | 0.00 |
ENSDART00000125923
|
cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr13_-_3022131 | 0.00 |
ENSDART00000050934
|
pkdcca
|
protein kinase domain containing, cytoplasmic a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.1 | 0.5 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.1 | 0.3 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.2 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.1 | 0.4 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 0.2 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.1 | 0.3 | GO:0090104 | pancreatic epsilon cell differentiation(GO:0090104) |
| 0.1 | 0.2 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.3 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.0 | 0.2 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.2 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.0 | 0.2 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.0 | 0.1 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.0 | 1.6 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.2 | GO:0003379 | establishment of cell polarity involved in gastrulation cell migration(GO:0003379) |
| 0.0 | 0.4 | GO:1902284 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 0.3 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.4 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.2 | GO:1900045 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.3 | GO:0090309 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.2 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.1 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.1 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.3 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.0 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.0 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 0.0 | GO:0090281 | negative regulation of vascular permeability(GO:0043116) negative regulation of calcium ion import(GO:0090281) |
| 0.0 | 0.2 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.1 | 0.4 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.1 | 0.2 | GO:0098594 | mucin granule(GO:0098594) |
| 0.0 | 0.2 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.3 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.0 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.0 | GO:0070724 | BMP receptor complex(GO:0070724) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 1.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.0 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.0 | GO:0030620 | U2 snRNA binding(GO:0030620) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |