Project
DANIO-CODE
Navigation
Downloads
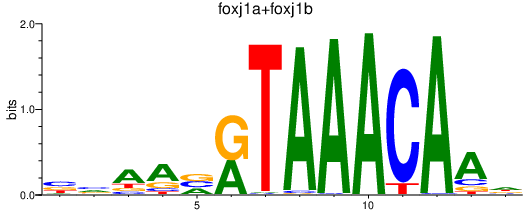
Results for foxj1a+foxj1b
Z-value: 0.98
Transcription factors associated with foxj1a+foxj1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxj1b
|
ENSDARG00000088290 | forkhead box J1b |
|
foxj1a
|
ENSDARG00000101919 | forkhead box J1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxj1a | dr10_dc_chr3_+_60637167_60637171 | -0.89 | 3.0e-06 | Click! |
Activity profile of foxj1a+foxj1b motif
Sorted Z-values of foxj1a+foxj1b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of foxj1a+foxj1b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_1562572 | 3.50 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr22_-_28828375 | 2.55 |
ENSDART00000104880
ENSDART00000005112 |
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr23_-_27123433 | 2.48 |
ENSDART00000142324
ENSDART00000133249 |
ENSDARG00000025766
|
ENSDARG00000025766 |
| chr1_-_7160294 | 2.48 |
ENSDART00000174972
|
BX908756.2
|
ENSDARG00000108483 |
| chr1_-_8593850 | 2.35 |
ENSDART00000146065
ENSDART00000114876 |
ubn1
|
ubinuclein 1 |
| chr4_-_20414881 | 2.16 |
ENSDART00000161343
|
cep83
|
centrosomal protein 83 |
| chr24_-_19573615 | 2.14 |
ENSDART00000158952
ENSDART00000109107 ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr17_-_23689233 | 2.03 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr5_+_61136434 | 1.94 |
ENSDART00000113508
|
hnrnpul1l
|
heterogeneous nuclear ribonucleoprotein U-like 1 like |
| chr1_-_7160099 | 1.88 |
ENSDART00000174972
|
BX908756.2
|
ENSDARG00000108483 |
| chr1_+_52686620 | 1.68 |
ENSDART00000176087
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr19_+_5399535 | 1.65 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr15_+_20054306 | 1.63 |
ENSDART00000155199
|
zgc:112083
|
zgc:112083 |
| chr9_+_41658126 | 1.60 |
ENSDART00000100265
|
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr13_-_25711537 | 1.47 |
ENSDART00000015154
|
papolg
|
poly(A) polymerase gamma |
| chr2_-_30216377 | 1.45 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr11_-_26819599 | 1.42 |
ENSDART00000043091
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr3_-_15325357 | 1.42 |
ENSDART00000139575
|
spns1
|
spinster homolog 1 (Drosophila) |
| chr1_+_52686770 | 1.42 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr13_+_29794944 | 1.38 |
ENSDART00000123482
|
cuedc2
|
CUE domain containing 2 |
| chr22_-_29740287 | 1.37 |
ENSDART00000166002
|
pdcd4b
|
programmed cell death 4b |
| chr15_+_34211736 | 1.34 |
ENSDART00000163513
|
arl4aa
|
ADP-ribosylation factor-like 4aa |
| chr4_+_16020464 | 1.33 |
ENSDART00000144611
|
CR749763.5
|
ENSDARG00000093983 |
| chr6_+_11162062 | 1.32 |
ENSDART00000151548
|
senp2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr20_+_48631807 | 1.32 |
ENSDART00000108884
|
prdm1c
|
PR domain containing 1c, with ZNF domain |
| chr21_+_10485148 | 1.31 |
ENSDART00000165070
|
ccbe1
|
collagen and calcium binding EGF domains 1 |
| chr15_-_14102102 | 1.30 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr2_+_1645259 | 1.29 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr14_-_32755189 | 1.26 |
ENSDART00000074720
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr15_+_27000467 | 1.25 |
ENSDART00000023842
|
ppm1da
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Da |
| chr23_-_1562603 | 1.23 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr13_+_29794969 | 1.20 |
ENSDART00000123482
|
cuedc2
|
CUE domain containing 2 |
| chr16_-_26947117 | 1.17 |
ENSDART00000111156
|
pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr15_-_28636418 | 1.17 |
ENSDART00000127845
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr17_+_14957568 | 1.17 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr2_-_30216333 | 1.16 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr13_-_31557441 | 1.16 |
ENSDART00000085989
|
trmt5
|
TRM5 tRNA methyltransferase 5 homolog (S. cerevisiae) |
| chr18_-_5167807 | 1.15 |
|
|
|
| chr7_-_25998808 | 1.15 |
ENSDART00000079347
|
serpine1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr7_+_36195803 | 1.15 |
ENSDART00000138893
|
aktip
|
akt interacting protein |
| chr2_-_17820860 | 1.12 |
ENSDART00000024302
|
kdm4ab
|
lysine (K)-specific demethylase 4A, genome duplicate b |
| chr6_+_40925259 | 1.11 |
ENSDART00000002728
ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr7_+_28453707 | 1.11 |
ENSDART00000173947
|
ccdc102a
|
coiled-coil domain containing 102A |
| chr22_-_29740737 | 1.11 |
ENSDART00000009223
|
pdcd4b
|
programmed cell death 4b |
| chr10_-_42279805 | 1.08 |
ENSDART00000132976
|
ess2
|
ess-2 splicing factor homolog |
| chr2_+_58943872 | 1.05 |
ENSDART00000158860
ENSDART00000067736 |
stk11
|
serine/threonine kinase 11 |
| chr13_+_31557497 | 1.04 |
ENSDART00000076479
|
slc38a6
|
solute carrier family 38, member 6 |
| chr14_+_14850200 | 1.04 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr10_-_3415830 | 1.03 |
ENSDART00000005168
|
ddx55
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 55 |
| chr10_-_14971273 | 1.02 |
ENSDART00000137430
|
smad2
|
SMAD family member 2 |
| chr9_+_8919663 | 1.01 |
ENSDART00000134954
|
carkd
|
carbohydrate kinase domain containing |
| chr23_-_27123338 | 1.00 |
ENSDART00000142324
ENSDART00000133249 |
ENSDARG00000025766
|
ENSDARG00000025766 |
| chr9_+_41658313 | 1.00 |
ENSDART00000100265
|
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr17_-_23689380 | 0.98 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr2_+_17027403 | 0.98 |
ENSDART00000164329
|
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr22_+_23260371 | 0.96 |
|
|
|
| chr11_+_16017857 | 0.96 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr22_+_23260413 | 0.96 |
|
|
|
| chr7_-_23840878 | 0.96 |
ENSDART00000111923
|
ajuba
|
ajuba LIM protein |
| chr20_+_53563389 | 0.95 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr9_-_2522639 | 0.94 |
ENSDART00000137706
|
scrn3
|
secernin 3 |
| chr1_+_50893368 | 0.94 |
ENSDART00000152789
|
etaa1
|
ETAA1, ATR kinase activator |
| chr19_-_24551474 | 0.94 |
ENSDART00000104087
|
thap7
|
THAP domain containing 7 |
| chr11_+_11136919 | 0.93 |
ENSDART00000026135
|
ly75
|
lymphocyte antigen 75 |
| chr23_-_36349563 | 0.93 |
|
|
|
| chr5_-_56254482 | 0.92 |
ENSDART00000014028
|
ppm1db
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Db |
| chr20_+_52968883 | 0.91 |
ENSDART00000014606
|
phactr1
|
phosphatase and actin regulator 1 |
| chr14_-_32755454 | 0.91 |
ENSDART00000074720
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr7_-_48532462 | 0.89 |
ENSDART00000015884
|
mfge8a
|
milk fat globule-EGF factor 8 protein a |
| chr22_+_18840143 | 0.89 |
|
|
|
| chr1_+_8468971 | 0.89 |
ENSDART00000081343
|
plk1
|
polo-like kinase 1 (Drosophila) |
| chr6_+_35811398 | 0.88 |
ENSDART00000151760
|
FP236809.1
|
ENSDARG00000096356 |
| chr14_-_6901209 | 0.87 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr14_+_27811681 | 0.86 |
|
|
|
| chr3_+_48810347 | 0.85 |
ENSDART00000133283
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr3_-_36298676 | 0.84 |
ENSDART00000157950
|
rogdi
|
rogdi homolog (Drosophila) |
| chr10_+_35313772 | 0.84 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr8_-_33145412 | 0.84 |
ENSDART00000133300
|
zbtb34
|
zinc finger and BTB domain containing 34 |
| chr10_-_42279667 | 0.83 |
ENSDART00000132976
|
ess2
|
ess-2 splicing factor homolog |
| chr20_+_48631724 | 0.83 |
ENSDART00000108884
|
prdm1c
|
PR domain containing 1c, with ZNF domain |
| chr7_+_36195907 | 0.82 |
ENSDART00000138893
|
aktip
|
akt interacting protein |
| chr10_-_35313462 | 0.80 |
ENSDART00000139107
|
prr11
|
proline rich 11 |
| chr20_+_52968725 | 0.80 |
ENSDART00000014606
|
phactr1
|
phosphatase and actin regulator 1 |
| chr24_+_24887139 | 0.80 |
ENSDART00000005845
|
mtmr6
|
myotubularin related protein 6 |
| chr11_-_26819525 | 0.80 |
ENSDART00000043091
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr16_+_26858575 | 0.78 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr5_+_32215942 | 0.78 |
ENSDART00000047377
|
crata
|
carnitine O-acetyltransferase a |
| chr17_+_32670368 | 0.78 |
ENSDART00000142449
|
ctsba
|
cathepsin Ba |
| chr17_-_23689317 | 0.77 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr19_-_5186692 | 0.75 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr3_+_33308831 | 0.75 |
ENSDART00000164322
|
gtpbp1
|
GTP binding protein 1 |
| chr14_-_32755370 | 0.75 |
ENSDART00000074720
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr1_+_52686829 | 0.74 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr6_-_41141242 | 0.74 |
ENSDART00000128723
ENSDART00000151055 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr14_+_8288174 | 0.74 |
ENSDART00000037749
|
stx5a
|
syntaxin 5A |
| chr8_-_28323975 | 0.74 |
|
|
|
| chr1_+_8469144 | 0.72 |
ENSDART00000081343
|
plk1
|
polo-like kinase 1 (Drosophila) |
| chr22_-_29740663 | 0.71 |
ENSDART00000009223
|
pdcd4b
|
programmed cell death 4b |
| chr23_+_24672435 | 0.71 |
ENSDART00000131689
ENSDART00000078796 |
kansl2
|
KAT8 regulatory NSL complex subunit 2 |
| chr7_-_56465369 | 0.71 |
ENSDART00000020967
|
csnk2a2a
|
casein kinase 2, alpha prime polypeptide a |
| chr18_+_2124267 | 0.71 |
|
|
|
| chr20_+_53563189 | 0.69 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr10_-_5135347 | 0.69 |
ENSDART00000138537
|
sec31a
|
SEC31 homolog A, COPII coat complex component |
| chr22_+_3167367 | 0.69 |
ENSDART00000160604
|
ftsj3
|
FtsJ homolog 3 (E. coli) |
| chr22_+_23260052 | 0.69 |
|
|
|
| chr7_+_38536733 | 0.68 |
ENSDART00000052323
|
harbi1
|
harbinger transposase derived 1 |
| chr24_+_14095601 | 0.68 |
ENSDART00000124740
|
ncoa2
|
nuclear receptor coactivator 2 |
| chr24_+_37752868 | 0.68 |
ENSDART00000158181
|
wdr24
|
WD repeat domain 24 |
| chr16_-_41764849 | 0.67 |
ENSDART00000084610
|
cep85
|
centrosomal protein 85 |
| chr19_+_5052459 | 0.66 |
ENSDART00000003634
|
stard3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr9_+_41657971 | 0.66 |
ENSDART00000132501
|
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr6_+_40924559 | 0.66 |
ENSDART00000133599
|
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr6_-_10492724 | 0.66 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr12_-_13927800 | 0.65 |
ENSDART00000066368
|
klhl11
|
kelch-like family member 11 |
| chr18_+_16759954 | 0.64 |
ENSDART00000061265
|
rnf141
|
ring finger protein 141 |
| chr17_-_20146897 | 0.63 |
ENSDART00000104874
|
p4ha1b
|
prolyl 4-hydroxylase, alpha polypeptide I b |
| chr12_+_43683149 | 0.62 |
ENSDART00000162014
|
si:ch211-232h12.2
|
si:ch211-232h12.2 |
| chr25_-_13774670 | 0.62 |
ENSDART00000160866
|
cry2
|
cryptochrome circadian clock 2 |
| chr3_-_45358502 | 0.62 |
ENSDART00000168108
|
prkacaa
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate a |
| chr23_+_44635770 | 0.61 |
|
|
|
| chr3_-_40522949 | 0.60 |
ENSDART00000178567
|
rnf216
|
ring finger protein 216 |
| chr22_-_28828300 | 0.59 |
ENSDART00000104880
|
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr5_+_36904127 | 0.59 |
ENSDART00000165465
|
si:ch1073-224n8.1
|
si:ch1073-224n8.1 |
| chr15_-_25693501 | 0.59 |
ENSDART00000155880
|
taok1a
|
TAO kinase 1a |
| chr5_+_61136494 | 0.58 |
ENSDART00000113508
|
hnrnpul1l
|
heterogeneous nuclear ribonucleoprotein U-like 1 like |
| chr1_-_25369230 | 0.58 |
ENSDART00000171752
|
pdcd4a
|
programmed cell death 4a |
| chr25_-_25047129 | 0.58 |
ENSDART00000031814
|
tsg101a
|
tumor susceptibility 101a |
| chr15_-_30935719 | 0.57 |
ENSDART00000050649
|
msi2b
|
musashi RNA-binding protein 2b |
| chr1_+_8594006 | 0.57 |
ENSDART00000142384
|
BX323038.1
|
ENSDARG00000092791 |
| chr15_-_28636615 | 0.57 |
ENSDART00000127845
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr2_-_10848705 | 0.56 |
ENSDART00000111298
|
mtf2
|
metal response element binding transcription factor 2 |
| KN150316v1_+_7339 | 0.55 |
|
|
|
| chr3_-_15325178 | 0.55 |
ENSDART00000139575
|
spns1
|
spinster homolog 1 (Drosophila) |
| chr5_-_39440181 | 0.55 |
ENSDART00000157755
|
cds1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr19_+_4939999 | 0.55 |
ENSDART00000093402
|
cdk12
|
cyclin-dependent kinase 12 |
| chr23_+_40070965 | 0.54 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr20_+_48631844 | 0.53 |
ENSDART00000108884
|
prdm1c
|
PR domain containing 1c, with ZNF domain |
| chr22_+_39012456 | 0.53 |
ENSDART00000176985
|
CABZ01044615.1
|
ENSDARG00000108810 |
| chr17_-_20147162 | 0.53 |
ENSDART00000104874
|
p4ha1b
|
prolyl 4-hydroxylase, alpha polypeptide I b |
| chr23_+_24672673 | 0.52 |
ENSDART00000131689
ENSDART00000078796 |
kansl2
|
KAT8 regulatory NSL complex subunit 2 |
| KN149858v1_+_13014 | 0.52 |
ENSDART00000165916
|
MED8
|
mediator complex subunit 8 |
| chr3_-_20643874 | 0.52 |
ENSDART00000163473
ENSDART00000159457 |
spop
|
speckle-type POZ protein |
| chr16_-_55263338 | 0.52 |
ENSDART00000113358
|
wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr2_-_30216703 | 0.50 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr25_-_34235582 | 0.50 |
ENSDART00000137665
|
frs2b
|
fibroblast growth factor receptor substrate 2b |
| chr6_-_41141280 | 0.50 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr20_-_18836698 | 0.50 |
ENSDART00000142837
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr10_+_42841562 | 0.50 |
|
|
|
| chr9_+_8920002 | 0.50 |
ENSDART00000132443
ENSDART00000139687 |
carkd
|
carbohydrate kinase domain containing |
| chr2_+_21208796 | 0.50 |
ENSDART00000062591
|
c2h1orf27
|
c2h1orf27 homolog (H. sapiens) |
| chr23_-_22144927 | 0.49 |
|
|
|
| chr23_+_7758140 | 0.49 |
ENSDART00000164255
|
pofut1
|
protein O-fucosyltransferase 1 |
| chr23_+_39569262 | 0.49 |
ENSDART00000031616
|
mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr3_+_54777037 | 0.48 |
ENSDART00000132587
|
mpg
|
N-methylpurine DNA glycosylase |
| chr25_-_34235633 | 0.47 |
ENSDART00000137665
|
frs2b
|
fibroblast growth factor receptor substrate 2b |
| chr18_-_35861412 | 0.47 |
ENSDART00000088488
|
opa3
|
optic atrophy 3 |
| chr23_-_30861356 | 0.46 |
ENSDART00000114628
|
myt1a
|
myelin transcription factor 1a |
| chr18_+_5260404 | 0.46 |
ENSDART00000150992
|
wdr59
|
WD repeat domain 59 |
| chr2_+_30976732 | 0.45 |
ENSDART00000141575
|
lipin2
|
lipin 2 |
| chr23_-_31339986 | 0.43 |
ENSDART00000139746
|
si:dkey-261l7.2
|
si:dkey-261l7.2 |
| chr15_+_27000248 | 0.42 |
ENSDART00000023842
|
ppm1da
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Da |
| KN149858v1_+_12702 | 0.42 |
ENSDART00000165916
|
MED8
|
mediator complex subunit 8 |
| chr2_-_22708324 | 0.42 |
|
|
|
| chr2_+_38288916 | 0.42 |
ENSDART00000042100
|
homeza
|
homeobox and leucine zipper encoding a |
| chr16_+_26858600 | 0.42 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr23_-_42983913 | 0.42 |
ENSDART00000148781
|
dlgap4a
|
discs, large (Drosophila) homolog-associated protein 4a |
| chr19_-_33013672 | 0.42 |
ENSDART00000134149
|
zgc:91944
|
zgc:91944 |
| chr14_+_38509121 | 0.41 |
ENSDART00000052511
|
hnrnpa0l
|
heterogeneous nuclear ribonucleoprotein A0, like |
| chr15_+_45738963 | 0.41 |
|
|
|
| chr8_-_1201388 | 0.41 |
ENSDART00000150064
|
cdc14b
|
cell division cycle 14B |
| chr5_+_44538762 | 0.41 |
ENSDART00000172702
ENSDART00000136002 |
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr19_-_10996809 | 0.41 |
ENSDART00000163179
|
pip5k1aa
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, a |
| chr2_-_51352799 | 0.41 |
ENSDART00000167987
|
ftr67
|
finTRIM family, member 67 |
| chr18_-_35861482 | 0.40 |
ENSDART00000088488
|
opa3
|
optic atrophy 3 |
| chr17_-_6294655 | 0.40 |
ENSDART00000064700
|
fuca2
|
fucosidase, alpha-L- 2, plasma |
| chr17_+_16600773 | 0.39 |
|
|
|
| chr15_-_14147062 | 0.39 |
ENSDART00000147796
|
trappc6bl
|
trafficking protein particle complex 6b-like |
| chr23_-_36350288 | 0.38 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
| chr22_+_132285 | 0.38 |
ENSDART00000059140
|
slc25a20
|
solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
| chr11_+_18450008 | 0.38 |
ENSDART00000162694
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr11_-_45042723 | 0.37 |
ENSDART00000163185
|
ankrd13c
|
ankyrin repeat domain 13C |
| chr15_+_24802607 | 0.37 |
ENSDART00000078024
|
crk
|
v-crk avian sarcoma virus CT10 oncogene homolog |
| chr7_-_38536517 | 0.37 |
ENSDART00000161397
|
atg13
|
ATG13 autophagy related 13 homolog (S. cerevisiae) |
| chr21_+_3645203 | 0.37 |
ENSDART00000099535
|
spout1
|
SPOUT domain containing methyltransferase 1 |
| chr3_-_36298866 | 0.37 |
ENSDART00000162875
|
rogdi
|
rogdi homolog (Drosophila) |
| chr7_-_50094750 | 0.36 |
ENSDART00000134941
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr3_-_36277758 | 0.36 |
ENSDART00000173545
|
cog1
|
component of oligomeric golgi complex 1 |
| chr10_+_23130414 | 0.35 |
ENSDART00000022170
|
LTN1
|
listerin E3 ubiquitin protein ligase 1 |
| chr20_+_16274045 | 0.35 |
ENSDART00000129633
|
zyg11
|
zyg-11 homolog (C. elegans) |
| chr12_-_22387786 | 0.35 |
ENSDART00000109707
|
neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr19_-_10997025 | 0.34 |
ENSDART00000163179
|
pip5k1aa
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, a |
| chr10_-_32946725 | 0.34 |
ENSDART00000134922
|
kctd7
|
potassium channel tetramerization domain containing 7 |
| chr8_-_39825792 | 0.34 |
ENSDART00000130686
|
unc119.1
|
unc-119 homolog 1 |
| chr24_-_25316192 | 0.34 |
ENSDART00000105820
|
mbtps2
|
membrane-bound transcription factor peptidase, site 2 |
| KN149858v1_+_12395 | 0.33 |
ENSDART00000165916
|
MED8
|
mediator complex subunit 8 |
| chr22_-_11490627 | 0.33 |
ENSDART00000063157
|
slc26a11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr2_+_41674839 | 0.32 |
ENSDART00000122860
ENSDART00000017977 |
acvr1l
|
activin A receptor, type I like |
| chr13_-_21519900 | 0.32 |
ENSDART00000078352
|
tspan14
|
tetraspanin 14 |
| chr10_-_35242344 | 0.31 |
ENSDART00000127805
|
pom121
|
POM121 transmembrane nucleoporin |
| chr22_+_23260220 | 0.31 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.8 | GO:0034036 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.4 | 1.3 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.4 | 1.2 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.3 | 1.2 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.3 | 0.8 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.3 | 1.3 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.3 | 0.8 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 0.9 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.2 | 2.0 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.2 | 0.8 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.2 | 3.6 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.2 | 1.3 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 0.5 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.2 | 0.5 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.1 | 1.0 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 1.2 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.1 | 2.0 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.1 | 1.0 | GO:2000637 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.1 | 0.3 | GO:0003179 | heart valve morphogenesis(GO:0003179) |
| 0.1 | 3.4 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.1 | 0.3 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 1.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 1.0 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 0.6 | GO:0071333 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.1 | 1.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 0.4 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.6 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 0.3 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.1 | 0.2 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.1 | 0.7 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.4 | GO:0000423 | macromitophagy(GO:0000423) |
| 0.1 | 0.4 | GO:0051657 | maintenance of organelle location(GO:0051657) |
| 0.1 | 2.5 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.1 | 0.6 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 1.5 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 1.7 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 1.0 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 6.0 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 2.3 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.5 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.4 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 0.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 2.0 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.8 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.7 | GO:0030301 | cholesterol transport(GO:0030301) |
| 0.0 | 0.9 | GO:0050879 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.0 | 0.2 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.6 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.3 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 0.3 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.9 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 1.3 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.9 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.8 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.6 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 1.2 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0090169 | regulation of spindle assembly(GO:0090169) regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 3.8 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 1.7 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.4 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.2 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.3 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.2 | GO:0090307 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.4 | 2.0 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 0.8 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 3.6 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.1 | 3.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 1.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.8 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.7 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 0.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.3 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.1 | 0.2 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.1 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.8 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 3.6 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.4 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.7 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.4 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 1.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.9 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.2 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.4 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.2 | GO:0045180 | cytoplasmic microtubule(GO:0005881) basal cortex(GO:0045180) |
| 0.0 | 1.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.2 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.4 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 1.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.5 | GO:0016459 | myosin complex(GO:0016459) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.8 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.4 | 1.5 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.3 | 0.8 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.3 | 0.8 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.2 | 1.2 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.2 | 1.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 1.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.2 | 0.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 1.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.6 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.1 | 2.8 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.1 | 0.6 | GO:0046922 | peptide-O-fucosyltransferase activity(GO:0046922) |
| 0.1 | 1.0 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 1.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.8 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 0.6 | GO:0004691 | AMP-activated protein kinase activity(GO:0004679) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.5 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.8 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 1.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 3.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.2 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 1.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 2.2 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.1 | 0.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.2 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.0 | 0.4 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.8 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.5 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.0 | 1.1 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.2 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) amidine-lyase activity(GO:0016842) |
| 0.0 | 1.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 1.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.0 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 3.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 3.0 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.0 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 0.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 2.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.2 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 0.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 2.7 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 0.5 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 3.1 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |