Project
DANIO-CODE
Navigation
Downloads
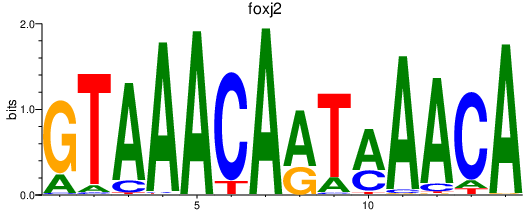
Results for foxj2
Z-value: 1.05
Transcription factors associated with foxj2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxj2
|
ENSDARG00000057680 | forkhead box J2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxj2 | dr10_dc_chr16_-_12896908_12896945 | 0.66 | 5.3e-03 | Click! |
Activity profile of foxj2 motif
Sorted Z-values of foxj2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of foxj2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_53286155 | 4.02 |
ENSDART00000139625
|
acta1a
|
actin, alpha 1a, skeletal muscle |
| chr13_+_3533517 | 2.77 |
ENSDART00000018737
|
qkib
|
QKI, KH domain containing, RNA binding b |
| chr18_+_5899355 | 2.68 |
|
|
|
| chr7_-_43501747 | 2.59 |
ENSDART00000002279
|
cdh11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr6_-_14821305 | 2.55 |
|
|
|
| chr15_-_18638643 | 2.42 |
ENSDART00000142010
|
ncam1b
|
neural cell adhesion molecule 1b |
| chr18_-_34166500 | 2.35 |
ENSDART00000079341
|
plch1
|
phospholipase C, eta 1 |
| chr16_+_14697969 | 2.34 |
ENSDART00000133566
|
deptor
|
DEP domain containing MTOR-interacting protein |
| chr4_-_19895115 | 2.27 |
ENSDART00000105967
|
cacna2d1a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1a |
| chr12_-_17357133 | 2.14 |
ENSDART00000079115
|
papss2b
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2b |
| chr16_+_24057260 | 2.10 |
ENSDART00000132742
ENSDART00000145330 |
apoc1
|
apolipoprotein C-I |
| chr1_-_19001718 | 2.06 |
ENSDART00000143303
|
BX323590.2
|
ENSDARG00000095388 |
| chr13_+_3533426 | 2.06 |
ENSDART00000018737
|
qkib
|
QKI, KH domain containing, RNA binding b |
| chr4_+_3344998 | 2.03 |
ENSDART00000075320
|
nampta
|
nicotinamide phosphoribosyltransferase a |
| chr5_-_23697487 | 1.98 |
ENSDART00000013309
|
sox19a
|
SRY (sex determining region Y)-box 19a |
| chr19_-_2486568 | 1.87 |
ENSDART00000012791
|
sp8a
|
sp8 transcription factor a |
| chr24_+_39213400 | 1.84 |
ENSDART00000115297
|
mss51
|
MSS51 mitochondrial translational activator |
| chr6_-_7281412 | 1.79 |
ENSDART00000053776
|
fkbp11
|
FK506 binding protein 11 |
| chr1_-_44476084 | 1.79 |
ENSDART00000034549
|
zgc:111983
|
zgc:111983 |
| chr25_+_5232865 | 1.74 |
|
|
|
| chr24_+_21395671 | 1.73 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr17_-_29102320 | 1.60 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr1_+_21037251 | 1.59 |
|
|
|
| chr7_+_28877570 | 1.57 |
ENSDART00000170342
|
mtbl
|
metallothionein-B-like |
| chr20_+_34867305 | 1.54 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr4_-_19895001 | 1.52 |
ENSDART00000014440
|
cacna2d1a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1a |
| chr11_-_22211478 | 1.51 |
ENSDART00000065996
|
tmem183a
|
transmembrane protein 183A |
| chr4_+_14728248 | 1.50 |
ENSDART00000124189
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr16_+_33189439 | 1.48 |
ENSDART00000101953
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr15_-_17933972 | 1.44 |
ENSDART00000155066
|
atf5b
|
activating transcription factor 5b |
| chr11_-_1792616 | 1.42 |
|
|
|
| chr15_-_18159904 | 1.37 |
ENSDART00000170874
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr3_-_57691670 | 1.34 |
ENSDART00000156522
|
cant1a
|
calcium activated nucleotidase 1a |
| chr7_-_28278286 | 1.33 |
ENSDART00000113313
|
st5
|
suppression of tumorigenicity 5 |
| chr22_-_6025108 | 1.31 |
|
|
|
| chr12_-_8780098 | 1.31 |
ENSDART00000113148
|
jmjd1cb
|
jumonji domain containing 1Cb |
| chr2_+_48448974 | 1.30 |
ENSDART00000023040
|
hes6
|
hes family bHLH transcription factor 6 |
| chr7_-_18448121 | 1.27 |
ENSDART00000112359
|
si:ch211-119e14.9
|
si:ch211-119e14.9 |
| chr3_-_6922655 | 1.27 |
|
|
|
| chr16_+_41922320 | 1.26 |
|
|
|
| chr8_+_23464087 | 1.24 |
ENSDART00000026316
|
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr5_-_8568715 | 1.23 |
ENSDART00000099891
|
atp5ib
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit Eb |
| chr6_+_7309023 | 1.23 |
ENSDART00000160128
|
erbb3a
|
erb-b2 receptor tyrosine kinase 3a |
| chr17_-_42288726 | 1.22 |
ENSDART00000154423
|
CR628331.1
|
ENSDARG00000097567 |
| chr12_-_9400344 | 1.21 |
ENSDART00000003932
|
erbb2
|
erb-b2 receptor tyrosine kinase 2 |
| chr3_-_6922811 | 1.21 |
|
|
|
| chr7_-_43501636 | 1.20 |
ENSDART00000002279
|
cdh11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr15_+_42617510 | 1.18 |
|
|
|
| chr10_+_23052796 | 1.16 |
ENSDART00000138955
|
si:dkey-175g6.2
|
si:dkey-175g6.2 |
| chr24_-_23178218 | 1.14 |
|
|
|
| chr17_-_37040941 | 1.11 |
ENSDART00000126823
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr5_-_70915664 | 1.09 |
|
|
|
| chr15_+_657554 | 1.06 |
ENSDART00000156007
|
si:ch73-144d13.8
|
si:ch73-144d13.8 |
| chr5_+_1223598 | 1.05 |
ENSDART00000159783
ENSDART00000026535 ENSDART00000168130 ENSDART00000147972 |
dnm1a
|
dynamin 1a |
| chr8_+_37667135 | 1.04 |
|
|
|
| chr14_-_25279835 | 1.04 |
ENSDART00000163669
|
cplx2
|
complexin 2 |
| chr5_+_45677439 | 1.03 |
ENSDART00000045598
|
zgc:110626
|
zgc:110626 |
| chr14_+_44312764 | 1.03 |
ENSDART00000098643
|
tomm5
|
translocase of outer mitochondrial membrane 5 homolog (yeast) |
| chr11_+_30569525 | 1.00 |
|
|
|
| chr8_+_47909655 | 0.98 |
|
|
|
| chr16_-_19154920 | 0.96 |
ENSDART00000088818
|
fhod3b
|
formin homology 2 domain containing 3b |
| chr6_-_29297541 | 0.95 |
ENSDART00000078630
|
nme7
|
NME/NM23 family member 7 |
| chr11_-_1374026 | 0.95 |
ENSDART00000172953
|
rpl29
|
ribosomal protein L29 |
| chr7_+_54410044 | 0.94 |
|
|
|
| chr17_-_1111115 | 0.93 |
|
|
|
| chr4_+_22020246 | 0.93 |
ENSDART00000112035
ENSDART00000127664 |
myf5
|
myogenic factor 5 |
| chr19_-_46058954 | 0.93 |
ENSDART00000163504
|
trps1
|
trichorhinophalangeal syndrome I |
| chr11_-_18305730 | 0.93 |
ENSDART00000155474
|
fgd5a
|
FYVE, RhoGEF and PH domain containing 5a |
| KN150178v1_-_23644 | 0.92 |
ENSDART00000165583
|
soul3
|
heme-binding protein soul3 |
| chr4_+_26368163 | 0.90 |
ENSDART00000148296
|
tnni1d
|
troponin I, skeletal, slow d |
| chr25_+_23326630 | 0.88 |
|
|
|
| chr9_+_6684835 | 0.88 |
|
|
|
| chr19_+_24398194 | 0.87 |
ENSDART00000080673
|
syt11a
|
synaptotagmin XIa |
| chr4_+_17364740 | 0.86 |
ENSDART00000136299
|
nup37
|
nucleoporin 37 |
| KN149921v1_-_848 | 0.85 |
|
|
|
| chr14_-_8584712 | 0.85 |
ENSDART00000167242
|
ATG2A
|
autophagy related 2A |
| chr1_-_44864706 | 0.84 |
|
|
|
| chr25_+_16593169 | 0.84 |
ENSDART00000073416
|
cecr1a
|
cat eye syndrome chromosome region, candidate 1a |
| chr3_+_28450576 | 0.83 |
ENSDART00000150893
|
sept12
|
septin 12 |
| chr3_+_13487523 | 0.83 |
ENSDART00000166000
|
si:ch211-194b1.1
|
si:ch211-194b1.1 |
| chr4_+_8796222 | 0.82 |
ENSDART00000158671
|
sult4a1
|
sulfotransferase family 4A, member 1 |
| chr2_-_24947660 | 0.82 |
ENSDART00000113356
ENSDART00000163038 |
crtc1a
|
CREB regulated transcription coactivator 1a |
| chr14_+_20614087 | 0.81 |
ENSDART00000160318
|
lygl2
|
lysozyme g-like 2 |
| chr24_-_38899781 | 0.80 |
ENSDART00000112318
|
ntn2
|
netrin 2 |
| chr23_+_32409339 | 0.79 |
ENSDART00000149698
|
slc39a5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr19_+_2162466 | 0.78 |
|
|
|
| KN150129v1_-_68034 | 0.77 |
ENSDART00000178130
|
CABZ01057446.1
|
ENSDARG00000106873 |
| chr7_+_69290855 | 0.77 |
ENSDART00000109507
|
RAP1GDS1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr25_+_28717079 | 0.75 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase, muscle, b |
| chr8_-_50299273 | 0.74 |
ENSDART00000023639
|
nkx2.7
|
NK2 transcription factor related 7 |
| chr3_+_6150498 | 0.73 |
ENSDART00000175915
|
BX284638.2
|
ENSDARG00000108888 |
| chr16_+_10427961 | 0.72 |
ENSDART00000055380
|
tubb5
|
tubulin, beta 5 |
| chr4_-_876235 | 0.71 |
|
|
|
| chr6_+_29297594 | 0.71 |
ENSDART00000112099
|
zgc:172121
|
zgc:172121 |
| chr24_-_27800234 | 0.69 |
ENSDART00000105778
|
hs6st1b
|
heparan sulfate 6-O-sulfotransferase 1b |
| chr2_-_39052185 | 0.69 |
ENSDART00000048838
|
rbp2b
|
retinol binding protein 2b, cellular |
| chr15_-_4536026 | 0.69 |
|
|
|
| chr10_-_11303185 | 0.69 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr10_-_13220838 | 0.68 |
ENSDART00000160265
|
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr18_+_38918956 | 0.68 |
ENSDART00000159834
ENSDART00000148541 ENSDART00000088037 |
myo5aa
|
myosin VAa |
| chr4_+_2415953 | 0.67 |
|
|
|
| chr19_+_20194594 | 0.66 |
ENSDART00000169074
|
hoxa4a
|
homeobox A4a |
| chr5_+_43365449 | 0.65 |
ENSDART00000113502
|
TMEM8B
|
si:dkey-84j12.1 |
| chr5_+_12090197 | 0.63 |
ENSDART00000133167
|
si:dkey-98f17.5
|
si:dkey-98f17.5 |
| chr20_-_26142897 | 0.61 |
ENSDART00000140255
|
si:dkey-12h9.6
|
si:dkey-12h9.6 |
| chr19_-_46058895 | 0.61 |
ENSDART00000163504
|
trps1
|
trichorhinophalangeal syndrome I |
| chr12_-_19224698 | 0.60 |
ENSDART00000044860
|
maff
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr6_-_43892838 | 0.60 |
ENSDART00000148646
ENSDART00000148909 |
foxp1b
|
forkhead box P1b |
| chr14_+_6684429 | 0.59 |
ENSDART00000157635
|
hnrnph1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr1_-_57842604 | 0.58 |
|
|
|
| chr16_+_24058139 | 0.58 |
|
|
|
| chr16_-_5254337 | 0.56 |
ENSDART00000134981
|
dctn3
|
dynactin 3 (p22) |
| chr19_-_48681377 | 0.56 |
ENSDART00000168065
|
si:ch73-359m17.5
|
si:ch73-359m17.5 |
| chr4_-_1989775 | 0.56 |
ENSDART00000003790
|
pwp1
|
PWP1 homolog (S. cerevisiae) |
| chr16_+_13928376 | 0.55 |
ENSDART00000163251
|
flcn
|
folliculin |
| chr5_-_68439806 | 0.54 |
ENSDART00000168213
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr5_+_61678044 | 0.53 |
ENSDART00000074117
|
aspa
|
aspartoacylase |
| chr14_-_46650329 | 0.51 |
|
|
|
| chr19_+_20194656 | 0.50 |
ENSDART00000169074
|
hoxa4a
|
homeobox A4a |
| chr8_+_17803229 | 0.50 |
ENSDART00000175436
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr14_-_1080574 | 0.49 |
ENSDART00000169090
ENSDART00000158905 ENSDART00000159907 |
ENSDARG00000098694
|
ENSDARG00000098694 |
| chr18_+_14725148 | 0.49 |
ENSDART00000146128
|
uri1
|
URI1, prefoldin-like chaperone |
| chr11_+_30416953 | 0.48 |
|
|
|
| chr14_-_28228583 | 0.48 |
ENSDART00000054088
|
zgc:113364
|
zgc:113364 |
| chr9_-_6402663 | 0.48 |
ENSDART00000078523
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr17_-_37040694 | 0.47 |
ENSDART00000126823
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr19_-_15324958 | 0.45 |
ENSDART00000055619
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr19_+_46631965 | 0.45 |
ENSDART00000158703
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr17_-_19606453 | 0.45 |
ENSDART00000011432
|
reep3a
|
receptor accessory protein 3a |
| chr17_+_34168095 | 0.44 |
|
|
|
| chr16_-_21064300 | 0.44 |
ENSDART00000040727
|
tax1bp1b
|
Tax1 (human T-cell leukemia virus type I) binding protein 1b |
| chr4_-_59767735 | 0.43 |
ENSDART00000159114
|
znf1129
|
zinc finger protein 1129 |
| chr4_+_9611685 | 0.42 |
ENSDART00000041289
|
tmem243b
|
transmembrane protein 243, mitochondrial b |
| chr3_+_59573755 | 0.41 |
ENSDART00000102014
|
fam195b
|
family with sequence similarity 195, member B |
| chr15_+_32863182 | 0.41 |
|
|
|
| chr14_-_46649718 | 0.41 |
|
|
|
| chr5_+_54642472 | 0.40 |
ENSDART00000131060
|
pcsk5a
|
proprotein convertase subtilisin/kexin type 5a |
| chr13_-_36193375 | 0.40 |
ENSDART00000057164
|
cox16
|
COX16 cytochrome c oxidase assembly homolog (S. cerevisiae) |
| chr1_+_50668743 | 0.39 |
ENSDART00000063938
|
mast1a
|
microtubule associated serine/threonine kinase 1a |
| chr2_-_11879298 | 0.38 |
ENSDART00000136748
|
ENSDARG00000039203
|
ENSDARG00000039203 |
| chr4_+_27111238 | 0.38 |
ENSDART00000141908
|
alg12
|
ALG12, alpha-1,6-mannosyltransferase |
| chr16_+_24057158 | 0.37 |
ENSDART00000132742
ENSDART00000145330 |
apoc1
|
apolipoprotein C-I |
| KN150170v1_-_8785 | 0.37 |
|
|
|
| chr23_+_40382464 | 0.37 |
ENSDART00000076876
|
fam46ab
|
family with sequence similarity 46, member Ab |
| chr7_+_9659464 | 0.36 |
ENSDART00000172824
|
cers3a
|
ceramide synthase 3a |
| chr12_-_8780030 | 0.34 |
ENSDART00000113148
|
jmjd1cb
|
jumonji domain containing 1Cb |
| chr1_-_22577723 | 0.34 |
ENSDART00000143948
|
rfc1
|
replication factor C (activator 1) 1 |
| chr17_+_16747316 | 0.33 |
ENSDART00000080129
|
ston2
|
stonin 2 |
| chr4_-_2415875 | 0.33 |
ENSDART00000125815
|
krr1
|
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr23_+_36648211 | 0.32 |
|
|
|
| chr18_-_14775081 | 0.32 |
ENSDART00000125979
|
tshz3a
|
teashirt zinc finger homeobox 3a |
| KN150030v1_-_22642 | 0.32 |
ENSDART00000175410
|
CABZ01079802.1
|
ENSDARG00000106760 |
| chr6_-_2037768 | 0.31 |
ENSDART00000159957
|
PXMP4
|
peroxisomal membrane protein 4 |
| chr17_-_29102271 | 0.30 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr23_-_17524325 | 0.30 |
ENSDART00000104680
|
tpd52l2b
|
tumor protein D52-like 2b |
| chr15_-_16140187 | 0.29 |
ENSDART00000080413
|
srsf1a
|
serine/arginine-rich splicing factor 1a |
| chr17_+_33462782 | 0.28 |
ENSDART00000140805
|
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr17_+_38655293 | 0.28 |
ENSDART00000062010
|
ccdc88c
|
coiled-coil domain containing 88C |
| chr14_-_46650465 | 0.27 |
|
|
|
| chr10_+_11303321 | 0.26 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr25_+_5233045 | 0.26 |
|
|
|
| chr20_+_34552759 | 0.25 |
ENSDART00000135789
|
mettl11b
|
methyltransferase like 11B |
| chr16_-_19154824 | 0.24 |
ENSDART00000088818
|
fhod3b
|
formin homology 2 domain containing 3b |
| chr3_-_28120760 | 0.23 |
|
|
|
| chr21_-_28629404 | 0.22 |
ENSDART00000125652
|
nrg2a
|
neuregulin 2a |
| chr22_+_1551406 | 0.22 |
ENSDART00000163223
|
si:ch211-255f4.9
|
si:ch211-255f4.9 |
| chr5_+_52362499 | 0.22 |
ENSDART00000157836
|
hint2
|
histidine triad nucleotide binding protein 2 |
| chr4_-_12796590 | 0.21 |
ENSDART00000075127
|
b2m
|
beta-2-microglobulin |
| chr7_+_35243375 | 0.20 |
ENSDART00000160065
|
BX294178.1
|
ENSDARG00000101818 |
| chr2_-_58214171 | 0.20 |
ENSDART00000169909
ENSDART00000168590 |
ENSDARG00000103284
|
ENSDARG00000103284 |
| chr18_+_38918883 | 0.20 |
ENSDART00000159202
|
myo5aa
|
myosin VAa |
| chr13_+_48565310 | 0.20 |
|
|
|
| chr14_-_46650551 | 0.19 |
|
|
|
| chr14_-_2451650 | 0.19 |
ENSDART00000111748
|
pcdhb
|
protocadherin b |
| chr13_+_46717255 | 0.19 |
ENSDART00000109247
|
anapc1
|
anaphase promoting complex subunit 1 |
| chr10_+_15106739 | 0.18 |
ENSDART00000128967
|
parm1
|
prostate androgen-regulated mucin-like protein 1 |
| chr10_+_26149600 | 0.18 |
|
|
|
| chr25_-_2208379 | 0.18 |
ENSDART00000168383
|
CR354430.4
|
ENSDARG00000101563 |
| chr4_-_21745163 | 0.17 |
ENSDART00000139962
|
pawr
|
PRKC, apoptosis, WT1, regulator |
| chr19_-_11928374 | 0.16 |
ENSDART00000148516
|
ctdp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr25_+_3091638 | 0.15 |
ENSDART00000104859
|
rccd1
|
RCC1 domain containing 1 |
| chr3_+_42212805 | 0.15 |
ENSDART00000169820
|
psmg3
|
proteasome (prosome, macropain) assembly chaperone 3 |
| chr5_-_67580755 | 0.15 |
ENSDART00000169350
|
ENSDARG00000098655
|
ENSDARG00000098655 |
| chr11_+_2641147 | 0.15 |
ENSDART00000132768
|
mapk14b
|
mitogen-activated protein kinase 14b |
| chr18_+_14665460 | 0.15 |
ENSDART00000133834
|
vps9d1
|
VPS9 domain containing 1 |
| chr10_+_29966533 | 0.14 |
ENSDART00000141549
|
hspa8
|
heat shock protein 8 |
| chr16_+_41922407 | 0.12 |
|
|
|
| chr22_-_12738507 | 0.12 |
ENSDART00000143574
|
rqcd1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr24_-_7558008 | 0.10 |
ENSDART00000093163
|
galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr18_+_41570495 | 0.10 |
ENSDART00000169621
ENSDART00000162052 |
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr9_+_2001849 | 0.10 |
ENSDART00000082329
|
evx2
|
even-skipped homeobox 2 |
| chr14_-_41751299 | 0.09 |
ENSDART00000128360
|
BX537109.1
|
ENSDARG00000086015 |
| chr24_+_25872051 | 0.07 |
ENSDART00000137851
|
tfr1b
|
transferrin receptor 1b |
| chr5_+_43365528 | 0.07 |
ENSDART00000113502
|
TMEM8B
|
si:dkey-84j12.1 |
| chr22_-_4715399 | 0.06 |
ENSDART00000122341
ENSDART00000161345 |
zfr2
|
zinc finger RNA binding protein 2 |
| chr23_-_18760927 | 0.06 |
ENSDART00000051182
|
arhgap4b
|
Rho GTPase activating protein 4b |
| chr3_-_61957965 | 0.05 |
ENSDART00000171952
ENSDART00000074174 |
tbl3
|
transducin (beta)-like 3 |
| chr8_-_1213205 | 0.04 |
ENSDART00000112130
|
aaed1
|
AhpC/TSA antioxidant enzyme domain containing 1 |
| chr10_+_3481047 | 0.03 |
ENSDART00000092684
|
rph3aa
|
rabphilin 3A homolog (mouse), a |
| chr8_+_31006910 | 0.02 |
ENSDART00000124213
|
odf2b
|
outer dense fiber of sperm tails 2b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.6 | 4.0 | GO:0072132 | mesenchyme morphogenesis(GO:0072132) |
| 0.4 | 1.2 | GO:0048940 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.4 | 2.1 | GO:0050428 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.3 | 1.6 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.3 | 0.8 | GO:0044036 | cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.3 | 3.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 0.9 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.2 | 1.5 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.2 | 1.0 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.2 | 1.9 | GO:0043584 | nose development(GO:0043584) |
| 0.2 | 0.6 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 1.7 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.0 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 0.7 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.1 | 0.8 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 1.5 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 1.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.8 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.7 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.1 | 0.7 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 1.8 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 0.2 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.1 | 2.3 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 0.8 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 0.5 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.5 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) regulation of TORC1 signaling(GO:1903432) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 0.7 | GO:0009086 | methionine metabolic process(GO:0006555) methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.9 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.4 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.2 | GO:0060347 | trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) |
| 0.0 | 1.6 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 4.7 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 1.0 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.5 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.0 | 0.1 | GO:1901741 | positive regulation of syncytium formation by plasma membrane fusion(GO:0060143) positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.2 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.4 | GO:0048899 | peptide hormone processing(GO:0016486) anterior lateral line development(GO:0048899) |
| 0.0 | 0.3 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.3 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 3.8 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 1.3 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 2.3 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 1.4 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 1.3 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 1.2 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.1 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.2 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.4 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.8 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.3 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 1.0 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 3.0 | GO:0006935 | chemotaxis(GO:0006935) |
| 0.0 | 1.4 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 1.3 | GO:0001756 | somitogenesis(GO:0001756) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.8 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.4 | 4.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 2.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 2.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 1.0 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 1.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 3.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.9 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.4 | GO:0045178 | basal part of cell(GO:0045178) basal cortex(GO:0045180) |
| 0.1 | 1.0 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.5 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 1.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 2.1 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.8 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 1.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.6 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 1.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.8 | GO:0005604 | basement membrane(GO:0005604) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.3 | 2.5 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.3 | 0.8 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.3 | 0.8 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 1.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.7 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 1.3 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 1.6 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 3.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 1.5 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 0.8 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.7 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 2.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 0.5 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 1.6 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.7 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 1.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.0 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 1.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 2.4 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 1.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 2.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.8 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 5.4 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 1.9 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 2.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.3 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 3.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.5 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 0.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.9 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 1.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |