Project
DANIO-CODE
Navigation
Downloads
Results for foxl1
Z-value: 1.32
Transcription factors associated with foxl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxl1
|
ENSDARG00000008133 | forkhead box L1 |
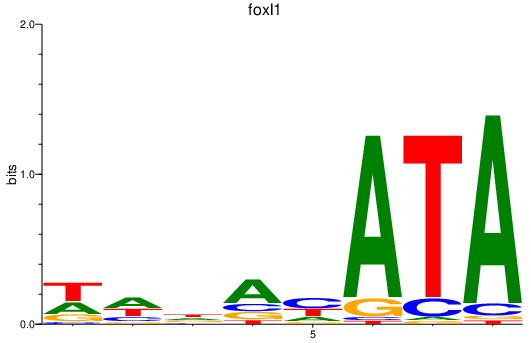
Activity profile of foxl1 motif
Sorted Z-values of foxl1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of foxl1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_19574944 | 6.65 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr13_+_2317444 | 4.37 |
ENSDART00000113692
|
tceb3l
|
transcription elongation factor B (SIII), polypeptide 3, like |
| chr25_-_9889107 | 3.18 |
ENSDART00000137407
|
AL929493.1
|
ENSDARG00000093575 |
| chr7_+_17695173 | 2.84 |
ENSDART00000101601
|
cth1
|
cysteine three histidine 1 |
| chr8_-_22493608 | 2.64 |
ENSDART00000021514
|
apex2
|
APEX nuclease (apurinic/apyrimidinic endonuclease) 2 |
| chr9_+_29709931 | 2.64 |
ENSDART00000078904
ENSDART00000150164 |
fdx1
|
ferredoxin 1 |
| chr13_-_6875290 | 2.58 |
|
|
|
| chr3_-_33296077 | 2.47 |
ENSDART00000075495
|
rpl23
|
ribosomal protein L23 |
| chr20_-_14218080 | 2.46 |
ENSDART00000104032
|
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr12_-_926596 | 2.38 |
ENSDART00000088351
|
SPAG9 (1 of many)
|
sperm associated antigen 9 |
| chr8_+_37716781 | 2.35 |
ENSDART00000108556
|
npm2a
|
nucleophosmin/nucleoplasmin, 2a |
| chr8_+_32380573 | 2.34 |
ENSDART00000146901
|
mthfd2l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr2_+_34984631 | 2.28 |
ENSDART00000141796
|
astn1
|
astrotactin 1 |
| chr19_-_35411993 | 2.20 |
ENSDART00000051751
|
zgc:113424
|
zgc:113424 |
| chr17_+_25313170 | 2.17 |
ENSDART00000157309
|
tmem54a
|
transmembrane protein 54a |
| chr17_+_1063519 | 2.04 |
|
|
|
| chr14_+_31278584 | 1.89 |
ENSDART00000158875
ENSDART00000026195 |
slc9a6a
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6a |
| chr18_-_6896957 | 1.82 |
ENSDART00000175747
|
ppp6r2b
|
protein phosphatase 6, regulatory subunit 2b |
| chr11_-_26819599 | 1.72 |
ENSDART00000043091
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr20_+_9487214 | 1.69 |
ENSDART00000053847
|
rad51b
|
RAD51 paralog B |
| chr20_+_14218237 | 1.63 |
ENSDART00000044937
|
kcns3b
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3b |
| chr6_+_23476669 | 1.60 |
|
|
|
| chr15_+_35076414 | 1.59 |
ENSDART00000165210
|
zgc:66024
|
zgc:66024 |
| chr17_-_24860924 | 1.58 |
ENSDART00000123147
|
zbtb8a
|
zinc finger and BTB domain containing 8A |
| chr20_-_14218236 | 1.57 |
ENSDART00000168434
|
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr14_-_46980330 | 1.52 |
ENSDART00000043751
ENSDART00000141357 |
macrod1
|
MACRO domain containing 1 |
| chr19_+_1298795 | 1.48 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr8_-_21110262 | 1.45 |
ENSDART00000143192
|
cpt2
|
carnitine palmitoyltransferase 2 |
| chr15_-_20189604 | 1.44 |
ENSDART00000152355
|
med13b
|
mediator complex subunit 13b |
| chr15_-_4976217 | 1.43 |
ENSDART00000101992
|
lipt2
|
lipoyl(octanoyl) transferase 2 (putative) |
| chr19_-_4206333 | 1.43 |
ENSDART00000164611
|
rragcb
|
Ras-related GTP binding Cb |
| chr5_+_64397858 | 1.42 |
ENSDART00000169209
|
ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr10_+_23090883 | 1.40 |
ENSDART00000079711
|
slc25a1a
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1a |
| chr10_+_32107014 | 1.40 |
ENSDART00000137373
|
si:ch211-266i6.3
|
si:ch211-266i6.3 |
| chr20_+_35535503 | 1.39 |
ENSDART00000153249
|
tdrd6
|
tudor domain containing 6 |
| chr7_+_36195907 | 1.39 |
ENSDART00000138893
|
aktip
|
akt interacting protein |
| chr4_-_13615927 | 1.35 |
ENSDART00000138366
|
irf5
|
interferon regulatory factor 5 |
| chr24_-_23639458 | 1.35 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr7_+_10319999 | 1.34 |
ENSDART00000168801
|
zfand6
|
zinc finger, AN1-type domain 6 |
| chr20_+_16740320 | 1.34 |
ENSDART00000152359
|
tmem30ab
|
transmembrane protein 30Ab |
| chr8_+_30103357 | 1.30 |
ENSDART00000133717
|
fancc
|
Fanconi anemia, complementation group C |
| chr9_-_33799681 | 1.30 |
ENSDART00000158678
|
caska
|
calcium/calmodulin-dependent serine protein kinase a |
| chr17_-_20182849 | 1.29 |
ENSDART00000133650
|
ecd
|
ecdysoneless homolog (Drosophila) |
| chr19_-_10411573 | 1.25 |
ENSDART00000171232
|
ccdc106b
|
coiled-coil domain containing 106b |
| chr1_+_13244109 | 1.25 |
ENSDART00000157563
|
noctb
|
nocturnin b |
| chr16_-_55211053 | 1.24 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr12_+_5067663 | 1.24 |
ENSDART00000166600
|
cep55l
|
centrosomal protein 55 like |
| chr13_-_24130160 | 1.22 |
ENSDART00000138747
ENSDART00000101168 |
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr11_-_17668381 | 1.21 |
ENSDART00000080752
|
uba3
|
ubiquitin-like modifier activating enzyme 3 |
| chr18_-_12889296 | 1.20 |
ENSDART00000130343
|
parp12a
|
poly (ADP-ribose) polymerase family, member 12a |
| chr7_-_55879342 | 1.20 |
ENSDART00000098438
|
spg7
|
spastic paraplegia 7 |
| chr13_+_11417882 | 1.19 |
ENSDART00000034935
|
desi2
|
desumoylating isopeptidase 2 |
| chr25_-_26399821 | 1.16 |
ENSDART00000140643
|
commd4
|
COMM domain containing 4 |
| chr20_-_50089784 | 1.14 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr3_+_14583287 | 1.14 |
|
|
|
| chr2_-_3115484 | 1.13 |
|
|
|
| chr5_+_22006830 | 1.12 |
ENSDART00000080919
|
rpl36a
|
ribosomal protein L36A |
| chr6_-_50731449 | 1.11 |
ENSDART00000157153
|
pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U |
| chr24_-_14447825 | 1.11 |
|
|
|
| chr23_-_26241101 | 1.11 |
|
|
|
| chr25_-_2932779 | 1.10 |
ENSDART00000149117
ENSDART00000137950 |
si:ch1073-296i8.2
|
si:ch1073-296i8.2 |
| chr7_-_58908421 | 1.08 |
ENSDART00000158996
|
nagk
|
N-acetylglucosamine kinase |
| chr7_+_29969850 | 1.08 |
ENSDART00000109243
|
sema4bb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Bb |
| chr12_-_18446876 | 1.08 |
ENSDART00000039693
|
pgp
|
phosphoglycolate phosphatase |
| chr5_+_22076136 | 1.08 |
ENSDART00000080877
|
tnfsf10l4
|
tumor necrosis factor (ligand) superfamily, member 10 like 4 |
| chr24_-_23639592 | 1.07 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr3_+_12087549 | 1.07 |
|
|
|
| chr13_+_29794944 | 1.06 |
ENSDART00000123482
|
cuedc2
|
CUE domain containing 2 |
| chr9_+_33523707 | 1.06 |
ENSDART00000171649
|
ddx3a
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3a |
| chr16_+_7463067 | 1.05 |
ENSDART00000149086
|
atg5
|
ATG5 autophagy related 5 homolog (S. cerevisiae) |
| KN150065v1_-_622 | 1.05 |
|
|
|
| chr14_-_14353451 | 1.04 |
ENSDART00000170355
ENSDART00000159888 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr12_-_37274632 | 1.04 |
ENSDART00000152951
|
cdc42ep4b
|
CDC42 effector protein (Rho GTPase binding) 4b |
| chr25_+_27300835 | 1.03 |
ENSDART00000103519
|
wasla
|
Wiskott-Aldrich syndrome-like a |
| chr5_-_23291887 | 1.03 |
ENSDART00000099083
ENSDART00000099084 ENSDART00000147887 |
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr3_+_27582277 | 1.03 |
ENSDART00000019004
|
arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr9_+_1820601 | 1.02 |
|
|
|
| chr15_-_47341913 | 1.02 |
ENSDART00000064462
|
psma6l
|
proteasome subunit alpha 6, like |
| chr25_-_13693524 | 1.02 |
ENSDART00000163512
ENSDART00000167742 |
ckap5
|
cytoskeleton associated protein 5 |
| chr22_-_21820673 | 1.01 |
ENSDART00000105564
|
aes
|
amino-terminal enhancer of split |
| chr23_+_7776647 | 1.01 |
ENSDART00000161193
|
kif3b
|
kinesin family member 3B |
| chr12_+_23939869 | 1.01 |
ENSDART00000021298
|
asb3
|
ankyrin repeat and SOCS box containing 3 |
| chr25_+_20174490 | 1.01 |
ENSDART00000109605
|
si:dkey-219c3.2
|
si:dkey-219c3.2 |
| chr24_+_26223767 | 1.01 |
|
|
|
| chr15_-_25158683 | 1.00 |
ENSDART00000129154
|
exo5
|
exonuclease 5 |
| chr6_-_50686517 | 0.99 |
ENSDART00000134146
|
mtss1
|
metastasis suppressor 1 |
| chr14_+_22170431 | 0.99 |
ENSDART00000079409
|
nudt22
|
nudix (nucleoside diphosphate linked moiety X)-type motif 22 |
| chr12_-_998693 | 0.98 |
ENSDART00000152346
|
polr3e
|
polymerase (RNA) III (DNA directed) polypeptide E |
| chr25_-_24150388 | 0.97 |
ENSDART00000073527
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr24_-_23639325 | 0.97 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr14_+_20941526 | 0.97 |
ENSDART00000138551
|
smim19
|
small integral membrane protein 19 |
| chr13_+_31557497 | 0.97 |
ENSDART00000076479
|
slc38a6
|
solute carrier family 38, member 6 |
| chr23_+_38875760 | 0.97 |
ENSDART00000086879
|
zfp64
|
zinc finger protein 64 homolog (mouse) |
| chr24_-_19574666 | 0.96 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr15_-_26954009 | 0.95 |
ENSDART00000156292
|
si:dkey-243i1.1
|
si:dkey-243i1.1 |
| chr8_-_21110183 | 0.95 |
ENSDART00000143192
|
cpt2
|
carnitine palmitoyltransferase 2 |
| chr11_-_3516034 | 0.95 |
ENSDART00000009788
|
ddx19
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 19 (DBP5 homolog, yeast) |
| chr7_+_52678718 | 0.95 |
ENSDART00000174216
|
cdkn2aip
|
CDKN2A interacting protein |
| chr6_-_6101185 | 0.95 |
ENSDART00000125918
ENSDART00000126142 ENSDART00000081966 |
rtn4a
|
reticulon 4a |
| chr20_-_34808532 | 0.92 |
ENSDART00000061555
|
si:ch211-63o20.7
|
si:ch211-63o20.7 |
| chr20_-_22899048 | 0.92 |
ENSDART00000063609
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr5_-_43205393 | 0.92 |
ENSDART00000028099
|
smn1
|
survival of motor neuron 1, telomeric |
| chr13_+_30993047 | 0.91 |
|
|
|
| chr10_+_36752015 | 0.91 |
ENSDART00000171392
|
rab6a
|
RAB6A, member RAS oncogene family |
| chr13_-_24614722 | 0.91 |
ENSDART00000031564
|
sfr1
|
SWI5-dependent homologous recombination repair protein 1 |
| chr1_-_54041101 | 0.91 |
ENSDART00000122601
|
pgam1b
|
phosphoglycerate mutase 1b |
| chr19_-_1571878 | 0.90 |
|
|
|
| chr21_-_17388020 | 0.89 |
ENSDART00000101295
|
gtf3c4
|
general transcription factor IIIC, polypeptide 4 |
| chr5_-_18393705 | 0.89 |
ENSDART00000010101
ENSDART00000099434 |
aacs
|
acetoacetyl-CoA synthetase |
| chr21_+_39378835 | 0.88 |
ENSDART00000031470
|
pafah1b1b
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1b |
| chr25_-_26399777 | 0.88 |
ENSDART00000140643
|
commd4
|
COMM domain containing 4 |
| chr2_-_24909432 | 0.87 |
ENSDART00000170283
|
crsp7
|
cofactor required for Sp1 transcriptional activation, subunit 7 |
| chr13_+_29794969 | 0.87 |
ENSDART00000123482
|
cuedc2
|
CUE domain containing 2 |
| chr18_+_18416922 | 0.87 |
ENSDART00000080174
|
n4bp1
|
nedd4 binding protein 1 |
| chr8_+_20108592 | 0.87 |
|
|
|
| chr8_+_26015379 | 0.86 |
ENSDART00000142555
|
arih2
|
ariadne homolog 2 (Drosophila) |
| chr22_-_31089831 | 0.86 |
|
|
|
| chr14_+_1451622 | 0.86 |
|
|
|
| chr11_-_26819525 | 0.85 |
ENSDART00000043091
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr18_+_22804237 | 0.85 |
ENSDART00000149685
|
gnao1a
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a |
| chr2_+_5383181 | 0.85 |
ENSDART00000019925
|
GNB4
|
G protein subunit beta 4 |
| chr24_+_39237314 | 0.83 |
ENSDART00000155346
|
tbc1d24
|
TBC1 domain family, member 24 |
| chr22_-_16416848 | 0.83 |
ENSDART00000006290
|
plekhb2
|
pleckstrin homology domain containing, family B (evectins) member 2 |
| chr2_+_5054170 | 0.82 |
ENSDART00000164626
|
si:ch211-162e15.3
|
si:ch211-162e15.3 |
| chr15_+_38319554 | 0.81 |
ENSDART00000122134
|
stim1a
|
stromal interaction molecule 1a |
| chr5_-_25570025 | 0.80 |
ENSDART00000017696
|
fam151b
|
family with sequence similarity 151, member B |
| chr19_-_31455278 | 0.80 |
ENSDART00000133101
ENSDART00000136213 |
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr6_-_52484653 | 0.80 |
ENSDART00000112146
|
fam83c
|
family with sequence similarity 83, member C |
| chr19_+_14295978 | 0.80 |
ENSDART00000168260
|
nudc
|
nudC nuclear distribution protein |
| chr2_+_5383347 | 0.79 |
ENSDART00000019925
|
GNB4
|
G protein subunit beta 4 |
| chr5_+_51186708 | 0.79 |
ENSDART00000165276
|
papd4
|
PAP associated domain containing 4 |
| chr4_+_4825461 | 0.79 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr20_+_9235730 | 0.79 |
ENSDART00000023293
|
kcnk5b
|
potassium channel, subfamily K, member 5b |
| chr24_+_14451260 | 0.79 |
ENSDART00000137337
ENSDART00000091784 |
thtpa
|
thiamine triphosphatase |
| chr15_-_16241412 | 0.78 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr20_-_33802082 | 0.78 |
ENSDART00000166573
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr7_-_71343686 | 0.78 |
ENSDART00000128678
|
mettl4
|
methyltransferase like 4 |
| chr12_+_4650974 | 0.78 |
ENSDART00000128145
|
kansl1a
|
KAT8 regulatory NSL complex subunit 1a |
| chr15_-_35008125 | 0.77 |
ENSDART00000099723
|
sh3bp5la
|
SH3-binding domain protein 5-like, a |
| chr17_+_39794317 | 0.77 |
ENSDART00000154996
|
si:dkey-229e3.2
|
si:dkey-229e3.2 |
| chr15_+_28242899 | 0.76 |
ENSDART00000037119
|
slc46a1
|
solute carrier family 46 (folate transporter), member 1 |
| chr24_-_10756995 | 0.76 |
ENSDART00000145593
|
fam49bb
|
family with sequence similarity 49, member Bb |
| chr18_+_20479090 | 0.76 |
ENSDART00000100665
ENSDART00000147867 |
ddb2
|
damage-specific DNA binding protein 2 |
| chr24_-_19574761 | 0.75 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr12_+_47828020 | 0.75 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr17_+_38499804 | 0.75 |
ENSDART00000149007
|
cdan1
|
codanin 1 |
| chr8_-_39788989 | 0.74 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr11_+_5724343 | 0.73 |
ENSDART00000172911
|
r3hdm4
|
R3H domain containing 4 |
| chr21_-_4901827 | 0.73 |
ENSDART00000067733
|
zgc:77838
|
zgc:77838 |
| chr19_-_6925090 | 0.72 |
ENSDART00000081568
|
tcf19l
|
transcription factor 19 (SC1), like |
| chr24_+_23571680 | 0.71 |
ENSDART00000080332
|
cops5
|
COP9 signalosome subunit 5 |
| chr21_-_1618494 | 0.71 |
ENSDART00000124904
|
zgc:152948
|
zgc:152948 |
| chr22_+_26736632 | 0.71 |
|
|
|
| chr12_+_27139848 | 0.70 |
ENSDART00000133023
|
tmem106a
|
transmembrane protein 106A |
| chr9_+_42355786 | 0.70 |
ENSDART00000142888
|
lrrc3
|
leucine rich repeat containing 3 |
| chr25_+_2252667 | 0.70 |
ENSDART00000172905
|
zmp:0000000932
|
zmp:0000000932 |
| chr11_-_2343369 | 0.69 |
ENSDART00000167032
ENSDART00000129854 |
tp53rk
|
TP53 regulating kinase |
| chr23_+_1693691 | 0.68 |
ENSDART00000149357
ENSDART00000099024 |
rabggta
|
Rab geranylgeranyltransferase, alpha subunit |
| chr21_-_30132195 | 0.68 |
ENSDART00000130820
|
hbegfb
|
heparin-binding EGF-like growth factor b |
| chr5_+_36250811 | 0.68 |
ENSDART00000051186
|
nccrp1
|
non-specific cytotoxic cell receptor protein 1 |
| chr12_+_23939934 | 0.66 |
ENSDART00000021298
|
asb3
|
ankyrin repeat and SOCS box containing 3 |
| chr22_+_33180599 | 0.66 |
ENSDART00000004504
|
dag1
|
dystroglycan 1 |
| chr14_+_15778201 | 0.66 |
ENSDART00000168462
|
rnf103
|
ring finger protein 103 |
| chr7_+_58448909 | 0.66 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr15_-_16447990 | 0.66 |
ENSDART00000154504
|
fam222bb
|
family with sequence similarity 222, member Bb |
| chr17_+_15780156 | 0.65 |
ENSDART00000027667
ENSDART00000161637 |
rragd
|
ras-related GTP binding D |
| chr11_+_2615745 | 0.65 |
ENSDART00000160777
|
b3galt6
|
UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6 |
| chr25_-_12712699 | 0.64 |
ENSDART00000162750
|
ca5a
|
carbonic anhydrase Va |
| chr7_+_26730804 | 0.64 |
|
|
|
| chr17_+_15666420 | 0.64 |
ENSDART00000156726
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
| chr22_-_28276824 | 0.64 |
ENSDART00000147686
|
si:dkey-222p3.1
|
si:dkey-222p3.1 |
| chr25_+_20596490 | 0.64 |
ENSDART00000073648
|
kxd1
|
KxDL motif containing 1 |
| chr22_+_1894340 | 0.62 |
ENSDART00000164158
|
znf1156
|
zinc finger protein 1156 |
| chr23_-_29825098 | 0.60 |
ENSDART00000056865
|
ctnnbip1
|
catenin, beta interacting protein 1 |
| chr15_+_17164535 | 0.60 |
ENSDART00000155350
|
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr3_+_44328013 | 0.60 |
ENSDART00000166294
|
metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr6_-_55312069 | 0.60 |
ENSDART00000083679
ENSDART00000162117 |
ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr7_+_67206056 | 0.60 |
ENSDART00000162553
|
kars
|
lysyl-tRNA synthetase |
| chr13_+_28560025 | 0.60 |
ENSDART00000137475
ENSDART00000128246 |
polr1c
|
polymerase (RNA) I polypeptide C |
| chr6_-_6101371 | 0.59 |
ENSDART00000125918
ENSDART00000126142 ENSDART00000081966 |
rtn4a
|
reticulon 4a |
| chr7_-_71343519 | 0.58 |
ENSDART00000128678
|
mettl4
|
methyltransferase like 4 |
| chr5_+_64397918 | 0.57 |
ENSDART00000162409
|
ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr22_-_21820611 | 0.57 |
ENSDART00000105564
|
aes
|
amino-terminal enhancer of split |
| chr1_+_23778514 | 0.57 |
ENSDART00000176581
|
fam160a1a
|
family with sequence similarity 160, member A1a |
| chr4_+_70728870 | 0.57 |
ENSDART00000122809
|
si:ch211-161m3.2
|
si:ch211-161m3.2 |
| chr22_+_2231377 | 0.56 |
ENSDART00000143366
|
si:dkey-4c15.8
|
si:dkey-4c15.8 |
| chr3_-_60883616 | 0.56 |
ENSDART00000156978
|
aimp2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr5_-_70089522 | 0.56 |
|
|
|
| chr18_-_44365869 | 0.56 |
ENSDART00000166935
|
prdm10
|
PR domain containing 10 |
| chr12_+_38541294 | 0.56 |
ENSDART00000130371
|
kif19
|
kinesin family member 19 |
| chr22_-_337759 | 0.56 |
|
|
|
| chr12_+_10668405 | 0.55 |
ENSDART00000161986
ENSDART00000158227 |
top2a
|
topoisomerase (DNA) II alpha |
| chr10_+_3299541 | 0.55 |
ENSDART00000031121
|
ENSDARG00000021564
|
ENSDARG00000021564 |
| chr11_+_42310157 | 0.55 |
ENSDART00000085868
|
appl1
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 |
| chr7_+_20266209 | 0.55 |
ENSDART00000052916
|
ENSDARG00000036426
|
ENSDARG00000036426 |
| chr15_-_23594653 | 0.55 |
ENSDART00000152543
|
hmbsb
|
hydroxymethylbilane synthase, b |
| chr21_-_36768113 | 0.55 |
ENSDART00000018350
|
cnot8
|
CCR4-NOT transcription complex, subunit 8 |
| chr19_-_9044008 | 0.55 |
ENSDART00000104657
|
mrps21
|
mitochondrial ribosomal protein S21 |
| chr11_-_34314863 | 0.55 |
ENSDART00000133302
|
pfkfb4a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4a |
| chr22_+_38322821 | 0.55 |
ENSDART00000104498
|
rcor3
|
REST corepressor 3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.6 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.4 | 2.3 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.4 | 1.1 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.4 | 1.4 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.3 | 1.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.3 | 1.0 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.3 | 1.9 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 1.2 | GO:0010482 | keratinocyte development(GO:0003334) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.3 | 0.9 | GO:0090277 | positive regulation of peptide secretion(GO:0002793) positive regulation of insulin secretion(GO:0032024) positive regulation of hormone secretion(GO:0046887) positive regulation of peptide hormone secretion(GO:0090277) |
| 0.3 | 0.8 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.3 | 0.8 | GO:0042357 | thiamine metabolic process(GO:0006772) thiamine diphosphate metabolic process(GO:0042357) thiamine-containing compound metabolic process(GO:0042723) |
| 0.3 | 1.5 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 2.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.2 | 0.9 | GO:1904396 | regulation of neuromuscular junction development(GO:1904396) |
| 0.2 | 1.5 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.2 | 1.0 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 1.0 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.2 | 1.1 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.2 | 0.7 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.2 | 0.5 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.2 | 8.4 | GO:0030901 | midbrain development(GO:0030901) |
| 0.2 | 1.4 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.1 | 0.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 1.0 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.1 | 1.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.4 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 1.0 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 0.5 | GO:0040014 | regulation of multicellular organism growth(GO:0040014) |
| 0.1 | 0.4 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 0.4 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 1.4 | GO:0030719 | P granule organization(GO:0030719) |
| 0.1 | 1.5 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.6 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 2.4 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.7 | GO:0060118 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.1 | 0.4 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.5 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.1 | 0.2 | GO:0045117 | azole transport(GO:0045117) |
| 0.1 | 0.9 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 3.3 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.7 | GO:0097065 | anterior head development(GO:0097065) |
| 0.1 | 1.0 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 0.9 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.9 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 1.0 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.3 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.1 | 1.0 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.7 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.1 | 2.0 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 2.6 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.6 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.1 | 0.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.2 | GO:0009202 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) |
| 0.1 | 2.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 1.7 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 0.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.7 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.1 | 0.4 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 1.3 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 0.2 | GO:1902633 | 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate metabolic process(GO:1902633) 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process(GO:1902635) |
| 0.1 | 0.8 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 0.6 | GO:0051340 | regulation of ligase activity(GO:0051340) positive regulation of ligase activity(GO:0051351) |
| 0.1 | 0.8 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 0.5 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.9 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.0 | 0.7 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.1 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.0 | 1.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.4 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.9 | GO:0048639 | positive regulation of developmental growth(GO:0048639) |
| 0.0 | 0.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 1.4 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.8 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.6 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 1.2 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.0 | 0.5 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 1.2 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 1.6 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 1.0 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.9 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) negative regulation of protein modification by small protein conjugation or removal(GO:1903321) |
| 0.0 | 0.4 | GO:0021694 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.4 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.8 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.7 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.2 | GO:1902110 | positive regulation of mitochondrial membrane permeability(GO:0035794) mitochondrial outer membrane permeabilization(GO:0097345) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.0 | 0.5 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 3.1 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 1.2 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 1.6 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 1.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.2 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.4 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.2 | GO:0043650 | dicarboxylic acid biosynthetic process(GO:0043650) |
| 0.0 | 0.1 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.8 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0044319 | wound healing, spreading of cells(GO:0044319) epiboly involved in wound healing(GO:0090505) skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.6 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.9 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 3.4 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 1.0 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.5 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 1.1 | GO:0006096 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 | 0.5 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.9 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.7 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.4 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.1 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.5 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 1.8 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.9 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.3 | GO:0042073 | intraciliary transport(GO:0042073) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.5 | 1.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.3 | 0.9 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.3 | 2.1 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.3 | 1.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 0.7 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 0.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 1.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.9 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 1.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.4 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 0.9 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.3 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 0.7 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 1.0 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.5 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 1.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 0.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.4 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.5 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.9 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 2.6 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 1.0 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.5 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.8 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 2.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.5 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 1.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.9 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.2 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 2.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.4 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 5.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 1.3 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.5 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.4 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.7 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.1 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.5 | 1.5 | GO:0016415 | octanoyltransferase activity(GO:0016415) |
| 0.5 | 2.3 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.4 | 2.8 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.4 | 1.5 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.4 | 1.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.3 | 1.9 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.3 | 2.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.3 | 0.8 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.2 | 1.0 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.2 | 1.7 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.2 | 0.7 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 0.7 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.2 | 1.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 1.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 3.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.2 | 1.4 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 0.9 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 1.0 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 1.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.5 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.1 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 2.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.2 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.1 | 0.8 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 1.0 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 1.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 0.8 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.5 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.1 | 0.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 0.8 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.2 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.1 | 0.6 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.1 | 0.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 1.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 0.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 1.0 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.4 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 1.4 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 0.2 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 1.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.4 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.1 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 2.1 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 0.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 1.0 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 1.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.1 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.8 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 1.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 1.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.9 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 0.4 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 1.7 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.8 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.5 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.7 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.1 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 2.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.7 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 1.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.5 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 0.2 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 1.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 1.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 2.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.7 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.2 | 2.8 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.0 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.9 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 1.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 0.9 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.0 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 0.5 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.8 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 1.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.8 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.0 | 1.1 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 2.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.5 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.7 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |