Project
DANIO-CODE
Navigation
Downloads
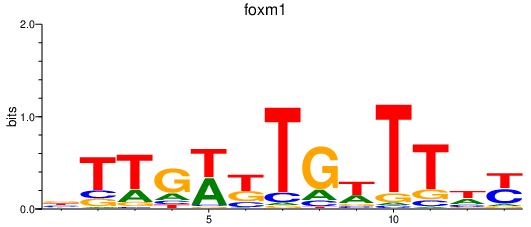
Results for foxm1
Z-value: 0.91
Transcription factors associated with foxm1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxm1
|
ENSDARG00000003200 | forkhead box M1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxm1 | dr10_dc_chr4_-_5822882_5823023 | 0.65 | 7.0e-03 | Click! |
Activity profile of foxm1 motif
Sorted Z-values of foxm1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of foxm1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_30013086 | 4.26 |
ENSDART00000138050
|
rbm33b
|
RNA binding motif protein 33b |
| KN150623v1_+_258 | 2.20 |
|
|
|
| chr18_-_11626694 | 2.08 |
ENSDART00000098565
|
CRACR2A
|
calcium release activated channel regulator 2A |
| chr17_-_45021393 | 2.04 |
|
|
|
| chr13_+_24549364 | 1.97 |
ENSDART00000139854
|
zgc:66426
|
zgc:66426 |
| chr18_+_17545564 | 1.73 |
ENSDART00000061007
|
mt2
|
metallothionein 2 |
| chr14_+_25208174 | 1.67 |
ENSDART00000079016
|
thoc3
|
THO complex 3 |
| chr22_+_818795 | 1.67 |
ENSDART00000003325
|
dennd2db
|
DENN/MADD domain containing 2Db |
| chr10_+_19059739 | 1.66 |
ENSDART00000038674
|
tmem230a
|
transmembrane protein 230a |
| chr1_-_50395003 | 1.58 |
ENSDART00000035150
|
spast
|
spastin |
| chr3_-_27470582 | 1.57 |
ENSDART00000077734
|
zgc:66160
|
zgc:66160 |
| chr10_-_4641624 | 1.55 |
ENSDART00000163951
|
plppr1
|
phospholipid phosphatase related 1 |
| chr15_-_29393378 | 1.54 |
ENSDART00000114492
|
si:dkey-52l18.4
|
si:dkey-52l18.4 |
| chr1_+_51118765 | 1.49 |
ENSDART00000156349
|
BX548032.3
|
ENSDARG00000096092 |
| chr4_+_16020464 | 1.30 |
ENSDART00000144611
|
CR749763.5
|
ENSDARG00000093983 |
| chr25_-_7794186 | 1.30 |
ENSDART00000104686
|
ambra1b
|
autophagy/beclin-1 regulator 1b |
| chr25_-_24105014 | 1.28 |
ENSDART00000048507
|
uevld
|
UEV and lactate/malate dehyrogenase domains |
| chr12_-_25288550 | 1.27 |
ENSDART00000142674
|
zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr3_-_54352535 | 1.23 |
ENSDART00000021977
ENSDART00000078973 |
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr7_-_2614300 | 1.21 |
ENSDART00000153548
|
BX323987.1
|
ENSDARG00000097745 |
| chr3_+_1398333 | 1.19 |
ENSDART00000031823
|
triobpb
|
TRIO and F-actin binding protein b |
| chr13_+_12629211 | 1.17 |
ENSDART00000015127
|
zgc:100846
|
zgc:100846 |
| chr17_-_7661725 | 1.17 |
ENSDART00000143870
|
rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr17_+_52526741 | 1.17 |
ENSDART00000109891
|
angel1
|
angel homolog 1 (Drosophila) |
| chr20_+_48631807 | 1.16 |
ENSDART00000108884
|
prdm1c
|
PR domain containing 1c, with ZNF domain |
| chr15_+_29460803 | 1.08 |
ENSDART00000155198
|
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr6_+_40924559 | 1.07 |
ENSDART00000133599
|
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr1_+_45148002 | 1.06 |
ENSDART00000148086
|
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr12_+_17032997 | 1.05 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr6_-_6951460 | 1.05 |
ENSDART00000148879
|
nhej1
|
nonhomologous end-joining factor 1 |
| chr22_+_1717236 | 1.04 |
|
|
|
| chr1_+_52686770 | 1.03 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr4_-_4603294 | 1.02 |
ENSDART00000130601
|
CABZ01020840.1
|
ENSDARG00000090401 |
| chr5_-_23092592 | 1.02 |
ENSDART00000024815
|
fam76b
|
family with sequence similarity 76, member B |
| chr8_-_22537326 | 1.01 |
ENSDART00000165640
|
porcnl
|
porcupine homolog like |
| chr6_+_40925259 | 1.01 |
ENSDART00000002728
ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr13_-_22831005 | 0.99 |
ENSDART00000143112
|
tspan15
|
tetraspanin 15 |
| chr25_-_28630138 | 0.98 |
|
|
|
| chr1_+_52686620 | 0.97 |
ENSDART00000176087
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr16_-_28659489 | 0.97 |
ENSDART00000059053
|
rpp38
|
ribonuclease P/MRP 38 subunit |
| chr13_-_22831037 | 0.96 |
ENSDART00000057641
|
tspan15
|
tetraspanin 15 |
| chr19_-_1571878 | 0.96 |
|
|
|
| chr14_-_15777250 | 0.95 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr2_+_30013537 | 0.94 |
ENSDART00000151848
|
rbm33b
|
RNA binding motif protein 33b |
| chr11_+_31346560 | 0.94 |
ENSDART00000124830
ENSDART00000162768 |
zgc:162816
|
zgc:162816 |
| chr4_+_9010972 | 0.94 |
ENSDART00000058007
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr3_-_22240424 | 0.92 |
|
|
|
| chr22_-_22107535 | 0.91 |
|
|
|
| chr4_+_14983045 | 0.90 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr20_+_26005068 | 0.90 |
ENSDART00000016864
|
slc35f6
|
solute carrier family 35, member F6 |
| chr23_-_33811895 | 0.87 |
ENSDART00000131680
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr17_-_11312263 | 0.86 |
ENSDART00000091159
|
adpgk2
|
ADP-dependent glucokinase 2 |
| chr2_-_44924505 | 0.86 |
ENSDART00000113351
|
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr17_-_7661762 | 0.85 |
ENSDART00000135538
|
rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr11_-_10001506 | 0.84 |
ENSDART00000123359
|
nlgn1
|
neuroligin 1 |
| chr15_-_29393320 | 0.84 |
ENSDART00000114492
|
si:dkey-52l18.4
|
si:dkey-52l18.4 |
| chr20_+_48631724 | 0.83 |
ENSDART00000108884
|
prdm1c
|
PR domain containing 1c, with ZNF domain |
| chr4_+_20901498 | 0.81 |
|
|
|
| chr20_+_46837436 | 0.80 |
ENSDART00000145294
|
ENSDARG00000022260
|
ENSDARG00000022260 |
| chr17_-_25612341 | 0.80 |
ENSDART00000126201
ENSDART00000105503 |
rab3gap2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
| chr2_+_37855019 | 0.78 |
ENSDART00000113337
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr24_+_5863770 | 0.78 |
ENSDART00000161104
|
mastl
|
microtubule associated serine/threonine kinase-like |
| chr7_-_3986977 | 0.78 |
|
|
|
| chr9_-_21729953 | 0.76 |
|
|
|
| chr5_-_9443859 | 0.74 |
|
|
|
| chr9_+_21448920 | 0.73 |
ENSDART00000137024
|
ska3
|
spindle and kinetochore associated complex subunit 3 |
| chr21_-_7322856 | 0.72 |
ENSDART00000151543
ENSDART00000114982 |
f2rl1.2
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 2 |
| KN149908v1_+_8072 | 0.71 |
|
|
|
| chr25_-_7794262 | 0.70 |
ENSDART00000156761
|
ambra1b
|
autophagy/beclin-1 regulator 1b |
| chr4_-_30690750 | 0.70 |
ENSDART00000160757
|
CU302413.2
|
ENSDARG00000107562 |
| chr17_-_23689380 | 0.69 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr13_+_33545847 | 0.69 |
ENSDART00000023379
|
mgme1
|
mitochondrial genome maintenance exonuclease 1 |
| chr13_-_51617421 | 0.68 |
ENSDART00000121457
|
lbh
|
limb bud and heart development |
| chr8_+_23126036 | 0.68 |
ENSDART00000141175
|
gid8a
|
GID complex subunit 8 homolog a (S. cerevisiae) |
| chr22_+_24532479 | 0.67 |
ENSDART00000169847
|
wdr47b
|
WD repeat domain 47b |
| chr15_-_28654137 | 0.67 |
ENSDART00000156049
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr23_+_37124543 | 0.66 |
ENSDART00000155706
|
CR762407.2
|
ENSDARG00000097111 |
| chr17_+_12878081 | 0.66 |
ENSDART00000115208
|
fbxo33
|
F-box protein 33 |
| chr21_-_23009910 | 0.66 |
ENSDART00000016167
|
zw10
|
zw10 kinetochore protein |
| chr10_-_244746 | 0.64 |
ENSDART00000136551
|
klhl35
|
kelch-like family member 35 |
| chr16_+_40181425 | 0.64 |
ENSDART00000155421
|
cenpw
|
centromere protein W |
| chr11_-_36212977 | 0.64 |
ENSDART00000165203
|
usp48
|
ubiquitin specific peptidase 48 |
| chr5_-_56874877 | 0.63 |
|
|
|
| chr22_-_38947928 | 0.63 |
ENSDART00000008365
|
ncbp2
|
nuclear cap binding protein subunit 2 |
| chr20_+_35156812 | 0.63 |
|
|
|
| chr20_-_22899048 | 0.63 |
ENSDART00000063609
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr13_-_33077267 | 0.63 |
ENSDART00000146138
|
trip11
|
thyroid hormone receptor interactor 11 |
| chr17_-_25612397 | 0.63 |
ENSDART00000126201
ENSDART00000105503 |
rab3gap2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
| chr9_-_56423275 | 0.63 |
|
|
|
| chr3_+_16772351 | 0.62 |
ENSDART00000164895
|
atp6v0a1a
|
ATPase, H+ transporting, lysosomal V0 subunit a1a |
| chr12_-_18997267 | 0.62 |
ENSDART00000149180
|
aco2
|
aconitase 2, mitochondrial |
| chr25_+_3521763 | 0.61 |
ENSDART00000160017
|
CABZ01058261.1
|
ENSDARG00000099194 |
| chr6_+_60130210 | 0.61 |
ENSDART00000148557
|
aurka
|
aurora kinase A |
| chr6_+_27100350 | 0.61 |
ENSDART00000121558
|
atg4b
|
autophagy related 4B, cysteine peptidase |
| chr8_-_602656 | 0.60 |
|
|
|
| chr13_+_24549249 | 0.60 |
ENSDART00000113981
|
zgc:66426
|
zgc:66426 |
| chr19_-_38832985 | 0.60 |
|
|
|
| chr4_-_36838449 | 0.59 |
|
|
|
| chr13_-_17880164 | 0.58 |
ENSDART00000144813
|
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr10_-_35108268 | 0.58 |
ENSDART00000147805
|
supt20
|
SPT20 homolog, SAGA complex component |
| chr8_-_24194821 | 0.58 |
ENSDART00000177464
|
CT025875.1
|
ENSDARG00000106046 |
| chr1_+_52686829 | 0.58 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr23_+_39718769 | 0.58 |
ENSDART00000034690
|
otud3
|
OTU deubiquitinase 3 |
| chr15_-_37687982 | 0.57 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr12_+_30119447 | 0.57 |
ENSDART00000102081
|
afap1l2
|
actin filament associated protein 1-like 2 |
| chr16_+_51328478 | 0.57 |
ENSDART00000168162
|
FQ323156.1
|
ENSDARG00000100697 |
| chr7_+_55648832 | 0.57 |
ENSDART00000082780
|
ACSF3
|
acyl-CoA synthetase family member 3 |
| chr7_+_23752492 | 0.56 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr3_+_1398365 | 0.56 |
ENSDART00000031823
|
triobpb
|
TRIO and F-actin binding protein b |
| chr19_+_25569817 | 0.56 |
ENSDART00000163220
|
si:ch211-239d6.2
|
si:ch211-239d6.2 |
| chr16_+_8067699 | 0.55 |
ENSDART00000126041
|
ano10a
|
anoctamin 10a |
| chr2_+_33943680 | 0.54 |
ENSDART00000109849
|
kif2c
|
kinesin family member 2C |
| chr25_-_25339397 | 0.53 |
ENSDART00000171589
|
hrasa
|
v-Ha-ras Harvey rat sarcoma viral oncogene homolog a |
| chr22_-_38947987 | 0.53 |
ENSDART00000008365
|
ncbp2
|
nuclear cap binding protein subunit 2 |
| chr20_+_48631844 | 0.51 |
ENSDART00000108884
|
prdm1c
|
PR domain containing 1c, with ZNF domain |
| chr10_+_22921629 | 0.51 |
ENSDART00000057291
|
arrb2a
|
arrestin, beta 2a |
| chr7_-_5172270 | 0.51 |
ENSDART00000172963
|
FP236789.1
|
ENSDARG00000105406 |
| chr4_-_20119812 | 0.51 |
ENSDART00000100867
|
fam3c
|
family with sequence similarity 3, member C |
| chr22_-_9905160 | 0.50 |
ENSDART00000136404
|
si:dkey-253d23.11
|
si:dkey-253d23.11 |
| chr8_-_18176067 | 0.50 |
ENSDART00000114177
|
si:ch211-241d21.5
|
si:ch211-241d21.5 |
| chr2_-_37821146 | 0.49 |
ENSDART00000154124
|
nfatc4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
| chr2_+_43355846 | 0.48 |
ENSDART00000056402
|
pard3ab
|
par-3 family cell polarity regulator alpha, b |
| chr14_+_44343015 | 0.48 |
ENSDART00000079866
|
slc30a9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr1_-_54488896 | 0.47 |
ENSDART00000150430
|
pane1
|
proliferation associated nuclear element |
| chr1_+_16834347 | 0.46 |
ENSDART00000005593
ENSDART00000140076 |
casp3a
|
caspase 3, apoptosis-related cysteine peptidase a |
| chr7_+_28844711 | 0.46 |
ENSDART00000052345
|
tradd
|
tnfrsf1a-associated via death domain |
| chr1_-_28057759 | 0.45 |
ENSDART00000152589
|
timmdc1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr24_-_2386805 | 0.44 |
|
|
|
| chr14_+_8332180 | 0.43 |
ENSDART00000123506
|
si:dkeyp-115e12.6
|
si:dkeyp-115e12.6 |
| chr11_+_30006715 | 0.43 |
ENSDART00000157272
ENSDART00000003475 |
ppef1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr7_-_38540389 | 0.42 |
|
|
|
| chr8_+_28377022 | 0.42 |
ENSDART00000158788
|
klhl12
|
kelch-like family member 12 |
| chr4_-_2370333 | 0.42 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr25_-_13990944 | 0.41 |
ENSDART00000124140
|
zgc:101566
|
zgc:101566 |
| chr24_+_5863597 | 0.41 |
ENSDART00000161104
|
mastl
|
microtubule associated serine/threonine kinase-like |
| chr8_+_23125906 | 0.41 |
ENSDART00000131617
|
gid8a
|
GID complex subunit 8 homolog a (S. cerevisiae) |
| chr20_+_46837276 | 0.41 |
ENSDART00000145294
|
ENSDARG00000022260
|
ENSDARG00000022260 |
| chr13_-_51617449 | 0.41 |
ENSDART00000121457
|
lbh
|
limb bud and heart development |
| chr4_+_5332964 | 0.39 |
ENSDART00000123375
|
zgc:113263
|
zgc:113263 |
| chr8_-_51537679 | 0.39 |
ENSDART00000147742
|
fgfr1a
|
fibroblast growth factor receptor 1a |
| chr8_+_23126151 | 0.39 |
ENSDART00000146264
|
gid8a
|
GID complex subunit 8 homolog a (S. cerevisiae) |
| chr2_-_47827343 | 0.38 |
ENSDART00000056882
|
cul3a
|
cullin 3a |
| chr18_+_18450245 | 0.38 |
ENSDART00000043843
|
siah1
|
siah E3 ubiquitin protein ligase 1 |
| chr17_-_15021379 | 0.38 |
ENSDART00000012877
ENSDART00000109190 |
ero1a
|
endoplasmic reticulum oxidoreductase alpha |
| chr18_-_20605032 | 0.38 |
ENSDART00000134722
|
bcl2l13
|
BCL2-like 13 (apoptosis facilitator) |
| chr21_-_2211701 | 0.38 |
ENSDART00000175686
|
si:dkey-50i6.6
|
si:dkey-50i6.6 |
| chr5_+_32215942 | 0.38 |
ENSDART00000047377
|
crata
|
carnitine O-acetyltransferase a |
| chr8_+_40176716 | 0.37 |
ENSDART00000055861
|
rnf34a
|
ring finger protein 34a |
| chr8_+_2398268 | 0.37 |
ENSDART00000133938
ENSDART00000002764 |
polb
|
polymerase (DNA directed), beta |
| chr15_+_31709330 | 0.37 |
ENSDART00000159634
|
b3glcta
|
beta 3-glucosyltransferase a |
| chr10_+_573116 | 0.37 |
ENSDART00000129856
ENSDART00000110384 |
smad4a
|
SMAD family member 4a |
| chr7_+_60054043 | 0.36 |
ENSDART00000145201
|
ppp1r14bb
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Bb |
| chr15_-_14653696 | 0.36 |
ENSDART00000172195
|
adck4
|
aarF domain containing kinase 4 |
| chr8_+_50543161 | 0.36 |
ENSDART00000176614
ENSDART00000174435 |
PEBP4
|
phosphatidylethanolamine binding protein 4 |
| chr15_+_23598869 | 0.36 |
ENSDART00000152320
|
si:dkey-182i3.10
|
si:dkey-182i3.10 |
| chr15_-_23849212 | 0.36 |
ENSDART00000059354
|
rad1
|
RAD1 homolog (S. pombe) |
| chr11_-_17882817 | 0.35 |
ENSDART00000155443
|
qrich1
|
glutamine-rich 1 |
| chr7_-_7533548 | 0.35 |
ENSDART00000173376
|
intu
|
inturned planar cell polarity protein |
| chr8_+_44448437 | 0.35 |
ENSDART00000143807
|
mhc1laa
|
major histocompatibility complex class I LAA |
| chr5_-_51256304 | 0.35 |
ENSDART00000125535
|
mtx3
|
metaxin 3 |
| chr6_+_27100249 | 0.35 |
ENSDART00000121558
|
atg4b
|
autophagy related 4B, cysteine peptidase |
| chr7_-_13661739 | 0.33 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr25_-_20633069 | 0.33 |
ENSDART00000172170
|
si:ch211-127m7.2
|
si:ch211-127m7.2 |
| chr4_-_2814833 | 0.33 |
|
|
|
| chr5_+_22470177 | 0.33 |
ENSDART00000020434
|
brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr22_+_38948272 | 0.32 |
ENSDART00000085685
|
sirt7
|
sirtuin 7 |
| chr19_+_19917767 | 0.32 |
|
|
|
| chr1_-_22577855 | 0.32 |
ENSDART00000175685
|
rfc1
|
replication factor C (activator 1) 1 |
| chr19_-_12058542 | 0.32 |
ENSDART00000168308
|
si:ch73-49k18.1
|
si:ch73-49k18.1 |
| chr20_+_38955797 | 0.32 |
|
|
|
| chr6_-_13910679 | 0.31 |
ENSDART00000065361
|
etv5b
|
ets variant 5b |
| chr14_+_15982295 | 0.31 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr15_+_1688846 | 0.30 |
ENSDART00000021299
|
nmd3
|
NMD3 ribosome export adaptor |
| chr17_+_8642373 | 0.30 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr7_-_6209659 | 0.28 |
ENSDART00000173032
|
CU457819.4
|
Histone H3.2 |
| chr10_+_15496705 | 0.28 |
ENSDART00000129441
|
erbb2ip
|
erbb2 interacting protein |
| chr2_+_11422195 | 0.28 |
ENSDART00000019078
|
lhx8a
|
LIM homeobox 8a |
| chr16_-_53978295 | 0.27 |
ENSDART00000158047
|
ZNRF2
|
zinc and ring finger 2 |
| chr11_-_44738541 | 0.27 |
ENSDART00000168066
|
afmid
|
arylformamidase |
| chr13_+_12629243 | 0.27 |
ENSDART00000015127
|
zgc:100846
|
zgc:100846 |
| chr9_+_42744931 | 0.27 |
|
|
|
| chr9_+_21729882 | 0.26 |
ENSDART00000132211
|
arhgef7a
|
Rho guanine nucleotide exchange factor (GEF) 7a |
| chr19_+_19164500 | 0.26 |
ENSDART00000165728
|
vps52
|
vacuolar protein sorting 52 homolog (S. cerevisiae) |
| chr10_+_19059663 | 0.26 |
ENSDART00000038674
|
tmem230a
|
transmembrane protein 230a |
| chr11_-_17883113 | 0.26 |
ENSDART00000040171
|
qrich1
|
glutamine-rich 1 |
| chr7_+_38540519 | 0.26 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr25_+_35540123 | 0.25 |
ENSDART00000126326
|
rpgrip1l
|
RPGRIP1-like |
| chr10_-_4641512 | 0.25 |
ENSDART00000163951
|
plppr1
|
phospholipid phosphatase related 1 |
| chr15_+_31709362 | 0.25 |
ENSDART00000159634
|
b3glcta
|
beta 3-glucosyltransferase a |
| chr22_+_1540035 | 0.24 |
ENSDART00000159050
|
si:ch211-255f4.8
|
si:ch211-255f4.8 |
| chr9_-_55250643 | 0.24 |
ENSDART00000085693
|
gpm6bb
|
glycoprotein M6Bb |
| chr18_+_15964146 | 0.24 |
|
|
|
| chr14_-_44329331 | 0.23 |
ENSDART00000098640
|
grhpra
|
glyoxylate reductase/hydroxypyruvate reductase a |
| chr14_-_51764932 | 0.23 |
ENSDART00000158353
|
exosc3
|
exosome component 3 |
| chr11_-_42148337 | 0.23 |
ENSDART00000160704
|
slmapa
|
sarcolemma associated protein a |
| chr1_+_39849028 | 0.23 |
ENSDART00000135578
|
scoca
|
short coiled-coil protein a |
| chr7_+_38540586 | 0.23 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr14_-_41020814 | 0.23 |
ENSDART00000169247
|
cstf2
|
cleavage stimulation factor, 3' pre-RNA, subunit 2 |
| chr23_-_5188577 | 0.22 |
ENSDART00000123191
|
ube2t
|
ubiquitin-conjugating enzyme E2T (putative) |
| chr22_+_1717284 | 0.22 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0043243 | positive regulation of protein complex disassembly(GO:0043243) |
| 0.4 | 1.2 | GO:0006408 | snRNA export from nucleus(GO:0006408) regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.3 | 2.0 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.3 | 0.8 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.3 | 1.0 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.2 | 0.7 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.2 | 1.7 | GO:0090050 | positive regulation of blood vessel endothelial cell migration(GO:0043536) positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.2 | 1.4 | GO:0036268 | swimming(GO:0036268) |
| 0.2 | 0.7 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 1.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.2 | 0.9 | GO:0019478 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.2 | 0.7 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.2 | 1.1 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.2 | 0.9 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 0.6 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.2 | 0.5 | GO:0043393 | G-protein coupled receptor internalization(GO:0002031) regulation of protein binding(GO:0043393) |
| 0.2 | 0.8 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.2 | 0.5 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 1.4 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.5 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.6 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 1.0 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.2 | GO:0051589 | negative regulation of neurotransmitter transport(GO:0051589) |
| 0.1 | 0.7 | GO:0034035 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.6 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.9 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.4 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.1 | 0.4 | GO:2001271 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.9 | GO:0032387 | negative regulation of intracellular transport(GO:0032387) |
| 0.1 | 1.5 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.1 | 0.5 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 1.8 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 0.6 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.1 | 0.2 | GO:1900274 | positive regulation of phospholipase C activity(GO:0010863) skin morphogenesis(GO:0043589) regulation of phospholipase C activity(GO:1900274) |
| 0.1 | 0.3 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 0.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.4 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 1.7 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 0.4 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.2 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 1.5 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 0.7 | GO:0071173 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.2 | GO:0010660 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.0 | 0.8 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.6 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.8 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.3 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.3 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.4 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 1.8 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.3 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 2.0 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.2 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 2.6 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.5 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.4 | GO:0050906 | detection of stimulus involved in sensory perception(GO:0050906) |
| 0.0 | 0.3 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.3 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.6 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 2.1 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 2.0 | GO:1990778 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 0.4 | GO:0060173 | limb development(GO:0060173) |
| 0.0 | 0.7 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 1.1 | GO:0003401 | axis elongation(GO:0003401) |
| 0.0 | 0.9 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.4 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0033314 | mitotic G2 DNA damage checkpoint(GO:0007095) mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.4 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.7 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 2.6 | GO:0051493 | regulation of cytoskeleton organization(GO:0051493) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.2 | 1.7 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 1.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.2 | 1.1 | GO:0070187 | telosome(GO:0070187) |
| 0.2 | 1.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.7 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.6 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 1.2 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.1 | 0.6 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.2 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.4 | GO:0000803 | sex chromosome(GO:0000803) inactive sex chromosome(GO:0098577) |
| 0.0 | 0.6 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 1.5 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 1.6 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 1.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.2 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 1.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.3 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.4 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.6 | GO:0060076 | excitatory synapse(GO:0060076) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.3 | 1.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.3 | 0.9 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.2 | 1.0 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.2 | 0.7 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 0.6 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.2 | 0.8 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.4 | GO:0008465 | glycerate dehydrogenase activity(GO:0008465) |
| 0.1 | 0.4 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.1 | 0.6 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 0.7 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.8 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.7 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.1 | 1.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 1.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.3 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.1 | 0.9 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 0.6 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.4 | GO:0017130 | TFIIH-class transcription factor binding(GO:0001097) poly(C) RNA binding(GO:0017130) |
| 0.1 | 1.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 1.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.6 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.5 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.4 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 1.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 2.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.0 | 0.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.4 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 1.1 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 2.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 2.3 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 1.0 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 2.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.1 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.2 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.5 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.8 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |