Project
DANIO-CODE
Navigation
Downloads
Results for foxn1
Z-value: 1.11
Transcription factors associated with foxn1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxn1
|
ENSDARG00000011879 | forkhead box N1 |
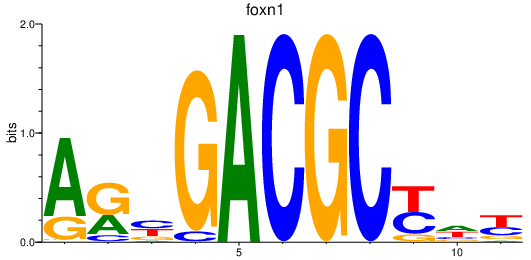
Activity profile of foxn1 motif
Sorted Z-values of foxn1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of foxn1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_19663746 | 3.48 |
ENSDART00000170149
|
slmapb
|
sarcolemma associated protein b |
| chr9_-_46614949 | 3.35 |
ENSDART00000009790
|
cx43.4
|
connexin 43.4 |
| chr17_-_45021393 | 2.93 |
|
|
|
| chr9_+_21982644 | 2.43 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr13_+_22973764 | 2.29 |
ENSDART00000110266
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr12_-_25058837 | 2.11 |
ENSDART00000135368
|
rhoq
|
ras homolog family member Q |
| KN150633v1_+_13413 | 2.04 |
|
|
|
| chr22_-_38473263 | 1.99 |
ENSDART00000098461
|
ptk7a
|
protein tyrosine kinase 7a |
| chr25_+_34340569 | 1.99 |
ENSDART00000157638
|
tmem231
|
transmembrane protein 231 |
| chr19_-_6274169 | 1.97 |
ENSDART00000140347
ENSDART00000092656 |
erf
|
Ets2 repressor factor |
| chr9_+_21982679 | 1.95 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr4_+_71390178 | 1.84 |
ENSDART00000174128
|
zgc:152938
|
zgc:152938 |
| chr8_+_5222023 | 1.80 |
ENSDART00000035676
|
bnip3la
|
BCL2/adenovirus E1B interacting protein 3-like a |
| chr6_+_40994859 | 1.78 |
ENSDART00000151419
|
tgfa
|
transforming growth factor, alpha |
| chr20_-_47576872 | 1.76 |
ENSDART00000067776
|
rab10
|
RAB10, member RAS oncogene family |
| chr13_-_50940551 | 1.73 |
|
|
|
| chr14_+_22159893 | 1.69 |
ENSDART00000019296
|
gdf9
|
growth differentiation factor 9 |
| chr6_+_40994918 | 1.67 |
ENSDART00000151419
|
tgfa
|
transforming growth factor, alpha |
| chr23_+_6652454 | 1.67 |
ENSDART00000081763
|
rbm38
|
RNA binding motif protein 38 |
| chr5_-_53907908 | 1.66 |
ENSDART00000158069
|
ssna1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr20_-_14885629 | 1.60 |
ENSDART00000160481
|
suco
|
SUN domain containing ossification factor |
| chr10_+_22065599 | 1.57 |
ENSDART00000143461
|
npm1a
|
nucleophosmin 1a (nucleolar phosphoprotein B23, numatrin) |
| chr25_-_36512943 | 1.56 |
ENSDART00000114508
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr4_-_12103129 | 1.53 |
ENSDART00000048391
ENSDART00000132650 |
braf
|
B-Raf proto-oncogene, serine/threonine kinase |
| chr5_+_29193876 | 1.53 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr20_-_14885599 | 1.53 |
ENSDART00000160481
|
suco
|
SUN domain containing ossification factor |
| chr3_+_59972297 | 1.52 |
|
|
|
| chr19_-_6274458 | 1.49 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr6_+_40995061 | 1.47 |
ENSDART00000151419
|
tgfa
|
transforming growth factor, alpha |
| chr13_-_50940520 | 1.46 |
|
|
|
| chr21_-_14154489 | 1.45 |
ENSDART00000114715
|
man1b1a
|
mannosidase, alpha, class 1B, member 1a |
| chr5_+_22541236 | 1.44 |
ENSDART00000171719
|
atrxl
|
alpha thalassemia/mental retardation syndrome X-linked, like |
| chr22_+_26840517 | 1.42 |
ENSDART00000158756
|
crebbpa
|
CREB binding protein a |
| chr23_+_30971404 | 1.40 |
ENSDART00000146859
|
cables2a
|
Cdk5 and Abl enzyme substrate 2a |
| chr10_+_29251234 | 1.40 |
ENSDART00000123033
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr9_+_21982756 | 1.35 |
ENSDART00000059652
|
rev1
|
REV1, polymerase (DNA directed) |
| chr22_+_26270259 | 1.33 |
ENSDART00000060898
|
mrps28
|
mitochondrial ribosomal protein S28 |
| chr5_-_29782745 | 1.33 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr8_-_602656 | 1.31 |
|
|
|
| chr10_-_1185831 | 1.30 |
ENSDART00000114261
|
bmpr1bb
|
bone morphogenetic protein receptor, type IBb |
| chr21_+_21242470 | 1.30 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr9_-_8916789 | 1.30 |
ENSDART00000144673
|
rab20
|
RAB20, member RAS oncogene family |
| chr22_-_38473305 | 1.29 |
ENSDART00000098461
|
ptk7a
|
protein tyrosine kinase 7a |
| chr3_-_15325357 | 1.28 |
ENSDART00000139575
|
spns1
|
spinster homolog 1 (Drosophila) |
| chr15_+_35105420 | 1.28 |
ENSDART00000153840
|
si:ch73-95l15.5
|
si:ch73-95l15.5 |
| chr22_+_62610 | 1.26 |
ENSDART00000062264
|
sdhb
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr13_+_35799681 | 1.24 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr13_+_35799602 | 1.24 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr5_-_18458903 | 1.24 |
ENSDART00000145210
|
ankle2
|
ankyrin repeat and LEM domain containing 2 |
| chr3_+_58417512 | 1.23 |
ENSDART00000154122
|
si:ch211-165g14.1
|
si:ch211-165g14.1 |
| chr3_+_1054368 | 1.22 |
ENSDART00000153693
|
si:ch73-166o21.1
|
si:ch73-166o21.1 |
| chr2_+_23004159 | 1.21 |
ENSDART00000123442
|
zgc:161973
|
zgc:161973 |
| chr3_+_18691103 | 1.20 |
ENSDART00000128626
|
tmem104
|
transmembrane protein 104 |
| chr2_-_32403565 | 1.20 |
ENSDART00000056634
|
ubtfl
|
upstream binding transcription factor, like |
| chr1_+_40428827 | 1.19 |
ENSDART00000145272
|
lrpap1
|
low density lipoprotein receptor-related protein associated protein 1 |
| chr3_-_18655432 | 1.18 |
ENSDART00000034489
|
msrb1a
|
methionine sulfoxide reductase B1a |
| chr16_+_26673760 | 1.17 |
ENSDART00000141393
|
ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr19_-_6274237 | 1.14 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr16_+_53632289 | 1.14 |
ENSDART00000124691
|
smpd5
|
sphingomyelin phosphodiesterase 5 |
| chr20_-_50211708 | 1.13 |
ENSDART00000148892
|
extl3
|
exostosin-like glycosyltransferase 3 |
| chr13_-_32324315 | 1.10 |
|
|
|
| chr7_+_66660882 | 1.08 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr13_+_28785104 | 1.07 |
|
|
|
| chr3_+_7877162 | 1.07 |
ENSDART00000057434
ENSDART00000170291 |
hook2
|
hook microtubule-tethering protein 2 |
| chr9_-_46615212 | 1.07 |
ENSDART00000009790
|
cx43.4
|
connexin 43.4 |
| chr9_-_41288163 | 1.05 |
ENSDART00000131681
|
asnsd1
|
asparagine synthetase domain containing 1 |
| chr5_-_34600655 | 1.05 |
ENSDART00000164396
|
fcho2
|
FCH domain only 2 |
| chr10_+_37556367 | 1.04 |
ENSDART00000135642
|
msi2a
|
musashi RNA-binding protein 2a |
| chr9_-_46615155 | 1.03 |
ENSDART00000009790
|
cx43.4
|
connexin 43.4 |
| chr17_+_34238753 | 1.02 |
ENSDART00000014306
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr19_+_29716509 | 1.02 |
ENSDART00000142694
ENSDART00000009149 |
tmem57a
|
transmembrane protein 57a |
| chr4_+_71390236 | 1.00 |
ENSDART00000174116
|
zgc:152938
|
zgc:152938 |
| chr13_+_32323781 | 1.00 |
ENSDART00000057421
|
rdh14a
|
retinol dehydrogenase 14a (all-trans/9-cis/11-cis) |
| chr3_-_29760674 | 0.99 |
ENSDART00000014021
|
slc25a39
|
solute carrier family 25, member 39 |
| chr16_-_40508858 | 0.99 |
ENSDART00000032389
|
plekhf2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr9_-_8916860 | 0.98 |
ENSDART00000138527
|
rab20
|
RAB20, member RAS oncogene family |
| chr10_+_29247498 | 0.98 |
|
|
|
| chr21_-_36710989 | 0.98 |
ENSDART00000086060
|
mrpl22
|
mitochondrial ribosomal protein L22 |
| chr9_-_8917143 | 0.98 |
ENSDART00000138527
|
rab20
|
RAB20, member RAS oncogene family |
| chr7_+_69290790 | 0.98 |
ENSDART00000109507
|
RAP1GDS1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr10_+_16543480 | 0.97 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr5_+_62595346 | 0.97 |
ENSDART00000140065
|
si:ch73-37h15.2
|
si:ch73-37h15.2 |
| chr10_+_384571 | 0.96 |
ENSDART00000129757
|
si:ch211-242f23.8
|
si:ch211-242f23.8 |
| chr22_-_38472936 | 0.95 |
ENSDART00000098461
|
ptk7a
|
protein tyrosine kinase 7a |
| chr3_-_29760552 | 0.94 |
ENSDART00000014021
|
slc25a39
|
solute carrier family 25, member 39 |
| chr19_+_29716557 | 0.94 |
ENSDART00000142694
ENSDART00000009149 |
tmem57a
|
transmembrane protein 57a |
| chr16_+_23367580 | 0.93 |
ENSDART00000015956
|
efna1b
|
ephrin-A1b |
| KN149779v1_-_14268 | 0.93 |
ENSDART00000162865
|
ift27
|
intraflagellar transport 27 homolog (Chlamydomonas) |
| chr23_+_35551931 | 0.92 |
ENSDART00000132609
|
ccnt1
|
cyclin T1 |
| chr2_-_48401770 | 0.91 |
ENSDART00000056277
|
gigfy2
|
GRB10 interacting GYF protein 2 |
| chr19_+_770644 | 0.91 |
ENSDART00000150926
|
gstr
|
glutathione S-transferase rho |
| chr8_+_5222145 | 0.91 |
ENSDART00000035676
|
bnip3la
|
BCL2/adenovirus E1B interacting protein 3-like a |
| chr25_-_34340016 | 0.90 |
ENSDART00000039485
|
gabarapl2
|
GABA(A) receptor-associated protein like 2 |
| chr6_-_7529384 | 0.90 |
ENSDART00000091836
ENSDART00000151697 |
ubn2a
|
ubinuclein 2a |
| chr20_+_23707789 | 0.90 |
|
|
|
| chr23_+_30971437 | 0.90 |
ENSDART00000146859
|
cables2a
|
Cdk5 and Abl enzyme substrate 2a |
| chr4_+_71390285 | 0.89 |
ENSDART00000174116
|
zgc:152938
|
zgc:152938 |
| chr17_-_52933930 | 0.88 |
|
|
|
| chr2_-_6380292 | 0.88 |
ENSDART00000092182
|
ppm1la
|
protein phosphatase, Mg2+/Mn2+ dependent, 1La |
| chr17_-_23874858 | 0.87 |
ENSDART00000122108
|
pdzd8
|
PDZ domain containing 8 |
| chr15_-_23973374 | 0.87 |
ENSDART00000088808
|
usp32
|
ubiquitin specific peptidase 32 |
| chr6_+_40994992 | 0.84 |
ENSDART00000151419
|
tgfa
|
transforming growth factor, alpha |
| chr10_+_29251158 | 0.83 |
ENSDART00000123033
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr18_+_18890626 | 0.83 |
ENSDART00000019581
|
arih1
|
ariadne ubiquitin-conjugating enzyme E2 binding protein homolog 1 (Drosophila) |
| chr7_+_8834138 | 0.82 |
ENSDART00000173250
|
pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr20_-_6153384 | 0.82 |
ENSDART00000161566
|
b4galt6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr11_-_22210506 | 0.81 |
ENSDART00000146873
|
tmem183a
|
transmembrane protein 183A |
| chr16_+_51597530 | 0.81 |
|
|
|
| chr8_+_13753663 | 0.80 |
|
|
|
| chr15_-_35020312 | 0.80 |
ENSDART00000154094
|
mgat1a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase a |
| chr5_+_1461715 | 0.80 |
ENSDART00000054057
|
ddrgk1
|
DDRGK domain containing 1 |
| chr20_+_23707625 | 0.79 |
|
|
|
| chr6_+_12269410 | 0.79 |
ENSDART00000149529
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr5_-_29782945 | 0.79 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr19_+_7254917 | 0.78 |
ENSDART00000123934
|
hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr8_-_21039978 | 0.78 |
ENSDART00000137606
ENSDART00000146532 |
zgc:112962
|
zgc:112962 |
| chr5_-_5848617 | 0.77 |
|
|
|
| chr5_+_34863545 | 0.77 |
ENSDART00000131286
|
erlin2
|
ER lipid raft associated 2 |
| chr24_+_19065679 | 0.77 |
ENSDART00000139299
|
zgc:162928
|
zgc:162928 |
| KN150034v1_+_1223 | 0.76 |
|
|
|
| chr5_-_29782972 | 0.76 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr7_+_23881828 | 0.75 |
ENSDART00000076735
|
lrp10
|
low density lipoprotein receptor-related protein 10 |
| chr7_-_600388 | 0.75 |
|
|
|
| chr1_+_43822319 | 0.74 |
ENSDART00000147702
|
zdhhc5a
|
zinc finger, DHHC-type containing 5a |
| chr8_+_52544578 | 0.74 |
ENSDART00000127729
|
stambpb
|
STAM binding protein b |
| chr6_+_50382153 | 0.74 |
ENSDART00000055504
|
cyc1
|
cytochrome c-1 |
| chr17_+_24045582 | 0.73 |
ENSDART00000132755
|
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr7_+_39812659 | 0.72 |
ENSDART00000099046
|
ENSDARG00000036489
|
ENSDARG00000036489 |
| chr3_-_19913207 | 0.72 |
|
|
|
| chr23_-_19560269 | 0.71 |
ENSDART00000009092
|
fam208ab
|
family with sequence similarity 208, member Ab |
| chr25_+_3423586 | 0.70 |
ENSDART00000166312
|
ccdc77
|
coiled-coil domain containing 77 |
| chr3_+_58417635 | 0.70 |
ENSDART00000154122
|
si:ch211-165g14.1
|
si:ch211-165g14.1 |
| chr13_+_22973830 | 0.70 |
ENSDART00000110266
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr2_-_16690351 | 0.68 |
ENSDART00000057216
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr4_+_26064174 | 0.68 |
ENSDART00000171204
|
SCYL2
|
SCY1 like pseudokinase 2 |
| chr17_-_16316519 | 0.68 |
ENSDART00000170091
|
hmbox1a
|
homeobox containing 1a |
| chr22_-_38472879 | 0.67 |
ENSDART00000149683
|
ptk7a
|
protein tyrosine kinase 7a |
| chr17_-_16316442 | 0.67 |
ENSDART00000170091
|
hmbox1a
|
homeobox containing 1a |
| chr5_+_25474996 | 0.67 |
ENSDART00000098473
|
klf9
|
Kruppel-like factor 9 |
| chr3_+_52745606 | 0.67 |
ENSDART00000104683
|
pbx4
|
pre-B-cell leukemia transcription factor 4 |
| chr5_-_18459312 | 0.67 |
ENSDART00000145210
|
ankle2
|
ankyrin repeat and LEM domain containing 2 |
| chr2_-_22875246 | 0.67 |
ENSDART00000159641
|
znf644a
|
zinc finger protein 644a |
| chr7_+_71585147 | 0.67 |
ENSDART00000161344
|
nsun6
|
NOL1/NOP2/Sun domain family, member 6 |
| chr4_+_71390404 | 0.66 |
ENSDART00000174116
|
zgc:152938
|
zgc:152938 |
| chr9_+_55395276 | 0.66 |
ENSDART00000029528
|
mospd2
|
motile sperm domain containing 2 |
| chr14_+_22160235 | 0.66 |
ENSDART00000019296
|
gdf9
|
growth differentiation factor 9 |
| chr1_-_44649941 | 0.66 |
ENSDART00000110390
|
ENSDARG00000025518
|
ENSDARG00000025518 |
| chr10_+_16543577 | 0.65 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr16_-_40508938 | 0.65 |
ENSDART00000032389
|
plekhf2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr9_-_28588192 | 0.64 |
ENSDART00000104317
ENSDART00000064343 |
klf7b
|
Kruppel-like factor 7b |
| chr13_-_25620394 | 0.64 |
ENSDART00000111567
|
sgpl1
|
sphingosine-1-phosphate lyase 1 |
| chr21_-_14079056 | 0.63 |
ENSDART00000111659
|
dfnb31a
|
deafness, autosomal recessive 31a |
| chr16_+_11351423 | 0.63 |
ENSDART00000138335
|
gsk3ab
|
glycogen synthase kinase 3 alpha b |
| chr17_+_43916865 | 0.63 |
ENSDART00000049503
|
peli2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr14_+_21457220 | 0.62 |
ENSDART00000111839
|
kdm2ab
|
lysine (K)-specific demethylase 2Ab |
| chr5_+_25475041 | 0.62 |
ENSDART00000098473
|
klf9
|
Kruppel-like factor 9 |
| chr7_+_23224924 | 0.62 |
ENSDART00000142401
|
zgc:109889
|
zgc:109889 |
| chr7_-_60046465 | 0.62 |
ENSDART00000127518
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr8_-_49217824 | 0.61 |
|
|
|
| chr3_+_15679209 | 0.61 |
ENSDART00000104397
|
tmem11
|
transmembrane protein 11 |
| chr11_-_37816180 | 0.61 |
ENSDART00000086516
|
klhdc8a
|
kelch domain containing 8A |
| chr17_-_23874694 | 0.61 |
ENSDART00000122108
|
pdzd8
|
PDZ domain containing 8 |
| chr25_+_7358777 | 0.61 |
ENSDART00000161593
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr5_-_18458827 | 0.60 |
ENSDART00000145210
|
ankle2
|
ankyrin repeat and LEM domain containing 2 |
| chr7_+_23224800 | 0.60 |
ENSDART00000115299
ENSDART00000101423 |
zgc:109889
|
zgc:109889 |
| chr17_-_32417510 | 0.60 |
ENSDART00000145487
|
klf11b
|
Kruppel-like factor 11b |
| chr8_-_25826973 | 0.60 |
ENSDART00000047008
ENSDART00000128829 |
efhd2
|
EF-hand domain family, member D2 |
| chr1_+_43822153 | 0.60 |
ENSDART00000147702
|
zdhhc5a
|
zinc finger, DHHC-type containing 5a |
| chr14_+_21522672 | 0.59 |
ENSDART00000122739
|
ctbp1
|
C-terminal binding protein 1 |
| chr8_+_16722505 | 0.58 |
ENSDART00000133514
|
elovl7a
|
ELOVL fatty acid elongase 7a |
| chr23_-_10851353 | 0.58 |
ENSDART00000055038
|
rybpa
|
RING1 and YY1 binding protein a |
| chr25_+_11212787 | 0.58 |
ENSDART00000159583
|
FQ312013.1
|
ENSDARG00000099473 |
| chr15_+_19399898 | 0.57 |
ENSDART00000143632
|
acad8
|
acyl-CoA dehydrogenase family, member 8 |
| chr22_-_6490288 | 0.57 |
ENSDART00000149676
|
CR352226.7
|
ENSDARG00000095952 |
| chr21_-_35291005 | 0.57 |
ENSDART00000134780
|
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr18_-_16948109 | 0.56 |
ENSDART00000100117
|
znf143b
|
zinc finger protein 143b |
| chr12_-_4597080 | 0.56 |
ENSDART00000110514
|
prr12a
|
proline rich 12a |
| chr25_-_16685648 | 0.56 |
ENSDART00000019413
|
ndufa9a
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9a |
| chr2_-_56632969 | 0.56 |
ENSDART00000089158
|
hmha1a
|
histocompatibility (minor) HA-1 a |
| chr18_+_439402 | 0.55 |
ENSDART00000167841
|
ss18l2
|
synovial sarcoma translocation gene on chromosome 18-like 2 |
| chr2_+_35750334 | 0.55 |
ENSDART00000052666
|
rasal2
|
RAS protein activator like 2 |
| chr13_-_25620444 | 0.55 |
ENSDART00000111567
|
sgpl1
|
sphingosine-1-phosphate lyase 1 |
| chr12_+_19199002 | 0.54 |
ENSDART00000066389
|
tmem184ba
|
transmembrane protein 184ba |
| chr9_-_28588228 | 0.53 |
ENSDART00000104317
ENSDART00000064343 |
klf7b
|
Kruppel-like factor 7b |
| chr7_-_60046429 | 0.52 |
ENSDART00000127518
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr7_+_39812732 | 0.52 |
ENSDART00000099046
|
ENSDARG00000036489
|
ENSDARG00000036489 |
| chr23_+_6652502 | 0.52 |
ENSDART00000081763
|
rbm38
|
RNA binding motif protein 38 |
| chr13_+_32323952 | 0.52 |
ENSDART00000057421
|
rdh14a
|
retinol dehydrogenase 14a (all-trans/9-cis/11-cis) |
| chr13_+_35799851 | 0.52 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr5_-_19428718 | 0.51 |
ENSDART00000088881
ENSDART00000168053 |
git2a
|
G protein-coupled receptor kinase interacting ArfGAP 2a |
| chr10_-_35164761 | 0.51 |
ENSDART00000049633
|
zgc:110006
|
zgc:110006 |
| chr4_-_16893974 | 0.51 |
ENSDART00000124627
|
strap
|
serine/threonine kinase receptor associated protein |
| chr14_+_21835107 | 0.51 |
ENSDART00000109065
|
ccng1
|
cyclin G1 |
| chr12_+_2769741 | 0.51 |
ENSDART00000178262
|
mms19
|
MMS19 homolog, cytosolic iron-sulfur assembly component |
| chr7_+_39812522 | 0.50 |
ENSDART00000099046
|
ENSDARG00000036489
|
ENSDARG00000036489 |
| chr17_+_24045786 | 0.50 |
ENSDART00000132755
|
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr9_-_55395115 | 0.50 |
|
|
|
| chr1_-_24453469 | 0.49 |
ENSDART00000132355
|
fbxw7
|
F-box and WD repeat domain containing 7 |
| chr15_-_35020250 | 0.49 |
ENSDART00000154094
|
mgat1a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase a |
| chr20_+_51918642 | 0.48 |
ENSDART00000010271
|
aida
|
axin interactor, dorsalization associated |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.9 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) |
| 0.8 | 5.7 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.8 | 3.0 | GO:0051977 | maintenance of blood-brain barrier(GO:0035633) lysophospholipid transport(GO:0051977) lipid transport across blood brain barrier(GO:1990379) |
| 0.7 | 5.8 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.5 | 1.5 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.4 | 1.8 | GO:0046098 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine metabolic process(GO:0046098) guanine biosynthetic process(GO:0046099) hypoxanthine metabolic process(GO:0046100) |
| 0.4 | 1.2 | GO:1902908 | regulation of melanosome transport(GO:1902908) |
| 0.4 | 2.3 | GO:0030237 | female sex determination(GO:0030237) |
| 0.4 | 3.1 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.4 | 1.2 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) negative regulation of calcium ion import(GO:0090281) |
| 0.4 | 1.9 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.3 | 1.4 | GO:0006407 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.3 | 2.5 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.3 | 1.2 | GO:0010985 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) regulation of lipoprotein particle clearance(GO:0010984) negative regulation of lipoprotein particle clearance(GO:0010985) |
| 0.3 | 0.8 | GO:1903894 | regulation of endoplasmic reticulum unfolded protein response(GO:1900101) negative regulation of endoplasmic reticulum unfolded protein response(GO:1900102) regulation of IRE1-mediated unfolded protein response(GO:1903894) negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.2 | 1.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 1.0 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.2 | 1.9 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.2 | 1.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.2 | 1.2 | GO:0017145 | stem cell division(GO:0017145) |
| 0.2 | 0.8 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.2 | 1.8 | GO:0097345 | positive regulation of mitochondrial membrane permeability(GO:0035794) mitochondrial outer membrane permeabilization(GO:0097345) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.2 | 0.6 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.5 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.1 | 0.5 | GO:0031642 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.1 | 0.8 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.3 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.1 | 1.0 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.9 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 1.2 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 1.3 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.1 | 1.2 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.1 | 1.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 1.4 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.8 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.7 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 0.5 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 1.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 1.5 | GO:0048538 | thymus development(GO:0048538) |
| 0.1 | 1.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 1.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.9 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.4 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 0.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.2 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.0 | 0.2 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.0 | 2.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 1.2 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.0 | 1.0 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.0 | 0.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 1.6 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.2 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.7 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.8 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.8 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 1.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.0 | 3.2 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.8 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.6 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.6 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.3 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.5 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 1.1 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.6 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.6 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.7 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.2 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.8 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.4 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.1 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 1.5 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 1.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 1.4 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.5 | GO:0039021 | pronephric glomerulus development(GO:0039021) |
| 0.0 | 0.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 3.5 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.7 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 0.1 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.3 | 1.5 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.2 | 0.5 | GO:0097361 | CIA complex(GO:0097361) |
| 0.2 | 0.8 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.2 | 0.8 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 1.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.7 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.9 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 0.2 | GO:0034991 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 1.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.7 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.2 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.0 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 1.0 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 3.3 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.2 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.0 | 0.5 | GO:0030663 | COPI vesicle coat(GO:0030126) COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 1.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.5 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 1.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.7 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.7 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 1.4 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 2.2 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 1.9 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 7.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.8 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 6.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.2 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 5.4 | GO:0005794 | Golgi apparatus(GO:0005794) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0017125 | deoxycytidyl transferase activity(GO:0017125) |
| 1.0 | 3.0 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.8 | 3.0 | GO:0051978 | lysophospholipid transporter activity(GO:0051978) |
| 0.6 | 1.8 | GO:0052657 | guanine phosphoribosyltransferase activity(GO:0052657) |
| 0.5 | 4.6 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.4 | 5.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.4 | 1.2 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.4 | 1.2 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.4 | 2.3 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.3 | 1.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.3 | 1.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.3 | 1.2 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.3 | 2.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 1.2 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.2 | 0.7 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.2 | 1.9 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.2 | 2.5 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.2 | 1.5 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.2 | 1.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 1.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 1.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 2.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.9 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 1.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 1.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 1.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.1 | 1.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.6 | GO:0022884 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.1 | 0.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.3 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.1 | 0.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 1.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.8 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.6 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 1.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 1.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.5 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 1.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.8 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.1 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.3 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.5 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.0 | 0.6 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 6.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.7 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 1.2 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 2.1 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.1 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 1.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 1.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 1.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 2.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 0.5 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 3.0 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.7 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.5 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 1.4 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.2 | 3.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 2.0 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 0.9 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 0.6 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 1.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 1.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 5.6 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 2.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.8 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |