Project
DANIO-CODE
Navigation
Downloads
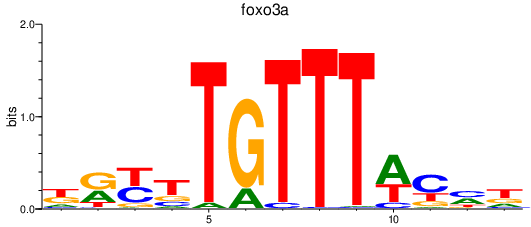
Results for foxo3a
Z-value: 0.43
Transcription factors associated with foxo3a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxo3a
|
ENSDARG00000023058 | forkhead box O3A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxo3a | dr10_dc_chr17_+_6635602_6635627 | -0.66 | 5.0e-03 | Click! |
Activity profile of foxo3a motif
Sorted Z-values of foxo3a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of foxo3a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_9992898 | 1.01 |
ENSDART00000080664
|
ACTC1
|
zgc:86709 |
| chr14_-_36037883 | 0.92 |
ENSDART00000173006
|
gpm6aa
|
glycoprotein M6Aa |
| chr8_+_7740132 | 0.81 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr25_-_30845998 | 0.73 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr6_-_14821305 | 0.72 |
|
|
|
| chr9_-_13383818 | 0.68 |
ENSDART00000084055
|
fzd7a
|
frizzled class receptor 7a |
| chr7_-_28425307 | 0.64 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr7_-_38288929 | 0.62 |
|
|
|
| chr4_-_6800721 | 0.53 |
ENSDART00000099467
|
ifrd1
|
interferon-related developmental regulator 1 |
| chr11_+_26371444 | 0.53 |
ENSDART00000042322
|
map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr21_+_10775209 | 0.52 |
|
|
|
| chr3_+_23567458 | 0.51 |
ENSDART00000078466
|
hoxb3a
|
homeobox B3a |
| chr8_-_24991984 | 0.51 |
ENSDART00000078795
|
ahcyl1
|
adenosylhomocysteinase-like 1 |
| chr9_+_25117850 | 0.50 |
|
|
|
| chr24_-_2316745 | 0.49 |
ENSDART00000138432
|
cul2
|
cullin 2 |
| chr15_-_25592228 | 0.49 |
ENSDART00000157498
|
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr3_+_23580269 | 0.46 |
ENSDART00000151584
|
hoxb4a
|
homeobox B4a |
| chr4_-_22751641 | 0.45 |
ENSDART00000066903
ENSDART00000130072 ENSDART00000123369 |
kmt2e
|
lysine (K)-specific methyltransferase 2E |
| chr12_+_16403743 | 0.45 |
ENSDART00000058665
|
kif20bb
|
kinesin family member 20Bb |
| chr7_-_28339836 | 0.44 |
ENSDART00000054366
|
scube2
|
signal peptide, CUB domain, EGF-like 2 |
| chr18_+_26916897 | 0.43 |
ENSDART00000050230
|
tspan3a
|
tetraspanin 3a |
| chr20_-_35568169 | 0.43 |
ENSDART00000153402
|
pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr11_+_6809190 | 0.43 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr16_-_1736470 | 0.42 |
ENSDART00000124660
|
ascc3
|
activating signal cointegrator 1 complex subunit 3 |
| chr11_-_1792616 | 0.40 |
|
|
|
| chr9_+_24089828 | 0.39 |
ENSDART00000127859
|
trim63b
|
tripartite motif containing 63b |
| chr13_+_24703802 | 0.39 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr7_+_47014195 | 0.38 |
ENSDART00000114669
|
dpy19l3
|
dpy-19-like 3 (C. elegans) |
| chr10_+_42761097 | 0.38 |
ENSDART00000075259
|
fgfr1b
|
fibroblast growth factor receptor 1b |
| chr15_-_4537178 | 0.38 |
ENSDART00000155619
ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr15_-_17074107 | 0.38 |
ENSDART00000156768
|
hip1
|
huntingtin interacting protein 1 |
| chr7_-_18295101 | 0.37 |
ENSDART00000173969
|
rgs12a
|
regulator of G protein signaling 12a |
| chr5_-_68439806 | 0.36 |
ENSDART00000168213
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr14_-_8975187 | 0.35 |
ENSDART00000054693
|
asb12b
|
ankyrin repeat and SOCS box-containing 12b |
| chr20_-_47680305 | 0.35 |
ENSDART00000023058
|
efhc1
|
EF-hand domain (C-terminal) containing 1 |
| chr16_-_42011213 | 0.35 |
ENSDART00000111956
|
gramd1a
|
GRAM domain containing 1A |
| chr2_+_9263844 | 0.35 |
ENSDART00000028906
|
pigk
|
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr25_+_13566205 | 0.35 |
ENSDART00000008989
|
ccdc113
|
coiled-coil domain containing 113 |
| chr15_-_17074348 | 0.34 |
ENSDART00000156768
|
hip1
|
huntingtin interacting protein 1 |
| chr6_+_7309023 | 0.34 |
ENSDART00000160128
|
erbb3a
|
erb-b2 receptor tyrosine kinase 3a |
| chr1_-_10423289 | 0.34 |
|
|
|
| chr18_-_38235232 | 0.32 |
ENSDART00000114224
|
si:dkey-10o6.2
|
si:dkey-10o6.2 |
| chr23_+_40070965 | 0.32 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr3_+_27477793 | 0.32 |
|
|
|
| chr23_+_33457343 | 0.31 |
ENSDART00000114423
|
rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr11_-_37921642 | 0.30 |
|
|
|
| chr7_-_28278286 | 0.30 |
ENSDART00000113313
|
st5
|
suppression of tumorigenicity 5 |
| chr23_-_4175790 | 0.30 |
ENSDART00000109807
|
ENSDARG00000076299
|
ENSDARG00000076299 |
| chr6_-_18733424 | 0.30 |
ENSDART00000151578
|
tns1a
|
tensin 1a |
| chr20_+_19175518 | 0.29 |
|
|
|
| chr25_-_3345176 | 0.28 |
ENSDART00000029067
|
hbp1
|
HMG-box transcription factor 1 |
| chr12_-_13848616 | 0.28 |
ENSDART00000110503
|
adam11
|
ADAM metallopeptidase domain 11 |
| chr6_-_9329917 | 0.27 |
ENSDART00000151470
|
map3k2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr25_+_2909562 | 0.26 |
ENSDART00000149360
|
mpi
|
mannose phosphate isomerase |
| chr4_-_14927871 | 0.26 |
ENSDART00000110199
|
prdm4
|
PR domain containing 4 |
| chr24_-_24859334 | 0.24 |
ENSDART00000080997
|
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr8_+_49582361 | 0.24 |
ENSDART00000108613
|
rasef
|
RAS and EF-hand domain containing |
| chr14_-_49951676 | 0.23 |
ENSDART00000021736
|
scocb
|
short coiled-coil protein b |
| chr14_+_32586048 | 0.23 |
ENSDART00000139159
|
lnx2b
|
ligand of numb-protein X 2b |
| chr1_-_5796394 | 0.23 |
ENSDART00000109356
|
klf7a
|
Kruppel-like factor 7a |
| chr13_+_35509853 | 0.23 |
ENSDART00000148481
|
thbs2a
|
thrombospondin 2a |
| chr9_-_10561062 | 0.23 |
ENSDART00000175269
|
thsd7ba
|
thrombospondin, type I, domain containing 7Ba |
| chr15_+_42393715 | 0.23 |
|
|
|
| chr5_-_54026450 | 0.23 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr10_-_36690119 | 0.22 |
ENSDART00000077161
|
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 |
| chr7_+_25778546 | 0.22 |
ENSDART00000173611
|
si:dkey-6n21.12
|
si:dkey-6n21.12 |
| chr17_+_44666606 | 0.22 |
ENSDART00000155096
|
tmem63c
|
transmembrane protein 63C |
| chr3_-_23580055 | 0.21 |
|
|
|
| chr15_+_25700036 | 0.21 |
ENSDART00000135409
ENSDART00000162240 |
tsr1
|
TSR1, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr8_-_3974357 | 0.21 |
ENSDART00000163754
ENSDART00000169474 |
mtmr3
|
myotubularin related protein 3 |
| chr23_-_12223878 | 0.21 |
ENSDART00000135035
|
fam217b
|
family with sequence similarity 217, member B |
| chr8_+_7739964 | 0.21 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr14_-_25301744 | 0.20 |
ENSDART00000131886
|
slc25a48
|
solute carrier family 25, member 48 |
| chr13_+_42476309 | 0.20 |
ENSDART00000133388
|
mlh1
|
mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) |
| chr12_+_34662699 | 0.19 |
ENSDART00000153097
|
slc38a10
|
solute carrier family 38, member 10 |
| chr15_+_30276801 | 0.18 |
ENSDART00000047248
ENSDART00000123937 |
nlk2
|
nemo-like kinase, type 2 |
| chr13_-_42274486 | 0.17 |
ENSDART00000043069
|
march5
|
membrane-associated ring finger (C3HC4) 5 |
| chr12_-_22387786 | 0.16 |
ENSDART00000109707
|
neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr1_-_29885008 | 0.16 |
ENSDART00000018827
|
dachc
|
dachshund c |
| chr25_-_10992022 | 0.15 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr21_-_22441376 | 0.14 |
|
|
|
| chr24_+_39314937 | 0.14 |
ENSDART00000129371
|
CABZ01072827.1
|
ENSDARG00000087471 |
| chr17_-_610651 | 0.14 |
|
|
|
| chr22_+_37696217 | 0.14 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr4_+_17364740 | 0.14 |
ENSDART00000136299
|
nup37
|
nucleoporin 37 |
| chr9_-_13383708 | 0.13 |
ENSDART00000084055
|
fzd7a
|
frizzled class receptor 7a |
| chr20_-_23526954 | 0.13 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr12_-_48278273 | 0.13 |
|
|
|
| chr3_+_23580216 | 0.13 |
ENSDART00000151584
|
hoxb4a
|
homeobox B4a |
| chr14_-_33704021 | 0.13 |
ENSDART00000149396
ENSDART00000123607 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr3_+_27655753 | 0.12 |
ENSDART00000086994
|
nat15
|
N-acetyltransferase 15 (GCN5-related, putative) |
| chr24_+_6649249 | 0.11 |
|
|
|
| chr23_-_33753634 | 0.11 |
ENSDART00000138416
|
tfcp2
|
transcription factor CP2 |
| chr10_+_29763134 | 0.11 |
|
|
|
| chr15_+_16961487 | 0.10 |
ENSDART00000154679
|
ypel2b
|
yippee-like 2b |
| chr3_+_52861409 | 0.10 |
ENSDART00000115117
|
brd4
|
bromodomain containing 4 |
| chr25_-_18153381 | 0.10 |
ENSDART00000067312
|
si:dkey-106n21.1
|
si:dkey-106n21.1 |
| chr12_-_19063851 | 0.10 |
ENSDART00000153343
|
zc3h7b
|
zinc finger CCCH-type containing 7B |
| chr17_-_26849495 | 0.09 |
ENSDART00000153590
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr25_-_35668179 | 0.09 |
ENSDART00000122559
ENSDART00000012944 |
amfr
|
autocrine motility factor receptor, E3 ubiquitin protein ligase a |
| chr21_-_41859818 | 0.08 |
ENSDART00000139698
|
endou2
|
endonuclease, polyU-specific 2 |
| chr13_+_24703728 | 0.08 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr5_-_4507692 | 0.08 |
ENSDART00000067600
|
zbtb43
|
zinc finger and BTB domain containing 43 |
| chr9_+_13258378 | 0.08 |
ENSDART00000141705
ENSDART00000154879 |
carf
|
calcium responsive transcription factor |
| chr5_-_51351827 | 0.07 |
ENSDART00000097194
|
serinc5
|
serine incorporator 5 |
| chr24_-_39749565 | 0.06 |
ENSDART00000142522
|
fam234a
|
family with sequence similarity 234, member A |
| chr10_-_6587490 | 0.06 |
ENSDART00000163788
ENSDART00000171833 |
chd1
|
chromodomain helicase DNA binding protein 1 |
| chr23_-_17524325 | 0.06 |
ENSDART00000104680
|
tpd52l2b
|
tumor protein D52-like 2b |
| chr25_-_7585373 | 0.06 |
ENSDART00000157076
|
prdm11
|
PR domain containing 11 |
| chr9_-_12913920 | 0.06 |
ENSDART00000122799
|
ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr12_+_27122352 | 0.05 |
ENSDART00000133048
|
nbr1a
|
neighbor of brca1 gene 1a |
| chr10_-_6587323 | 0.05 |
ENSDART00000163788
ENSDART00000171833 |
chd1
|
chromodomain helicase DNA binding protein 1 |
| chr6_-_54118302 | 0.05 |
ENSDART00000161059
|
tusc2a
|
tumor suppressor candidate 2a |
| chr24_-_32110851 | 0.04 |
ENSDART00000159034
|
rsu1
|
Ras suppressor protein 1 |
| chr9_+_13258490 | 0.04 |
ENSDART00000141705
|
carf
|
calcium responsive transcription factor |
| chr18_+_27119086 | 0.04 |
ENSDART00000086094
|
plekha7a
|
pleckstrin homology domain containing, family A member 7a |
| chr5_-_19942216 | 0.04 |
ENSDART00000051607
|
ISCU (1 of many)
|
iron-sulfur cluster assembly enzyme |
| chr14_-_32838287 | 0.04 |
|
|
|
| chr8_+_21192955 | 0.04 |
ENSDART00000129210
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr1_+_44113645 | 0.03 |
|
|
|
| chr16_+_36794641 | 0.03 |
ENSDART00000139069
|
decr1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr5_+_36010448 | 0.03 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr3_-_19913881 | 0.03 |
ENSDART00000126915
|
ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr10_-_8238422 | 0.03 |
ENSDART00000129467
|
dhx29
|
DEAH (Asp-Glu-Ala-His) box polypeptide 29 |
| chr7_-_32562435 | 0.03 |
ENSDART00000099872
ENSDART00000099871 ENSDART00000147554 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr18_-_40923017 | 0.02 |
ENSDART00000098878
|
polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr19_-_13045465 | 0.02 |
ENSDART00000128975
|
slc25a32a
|
solute carrier family 25 (mitochondrial folate carrier), member 32a |
| chr3_-_13396181 | 0.01 |
ENSDART00000159647
|
amdhd2
|
amidohydrolase domain containing 2 |
| chr1_+_45148094 | 0.01 |
ENSDART00000048191
|
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr24_-_24999348 | 0.01 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr25_+_26962541 | 0.01 |
ENSDART00000115139
|
pot1
|
protection of telomeres 1 homolog |
| chr5_-_66671163 | 0.00 |
ENSDART00000144092
|
mlxip
|
MLX interacting protein |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0070445 | peripheral nervous system myelin maintenance(GO:0032287) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.2 | 0.7 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.2 | 1.0 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.2 | 0.8 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 0.4 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.1 | 0.3 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.4 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.4 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 0.5 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.4 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.3 | GO:0071554 | cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) chondrocyte morphogenesis(GO:0090171) |
| 0.0 | 0.4 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.0 | 0.1 | GO:0051311 | chiasma assembly(GO:0051026) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.1 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.0 | 0.4 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.5 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.0 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.2 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.2 | GO:0051181 | cofactor transport(GO:0051181) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.5 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.5 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.5 | GO:0016801 | hydrolase activity, acting on ether bonds(GO:0016801) |
| 0.1 | 0.4 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 0.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.4 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.0 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 0.0 | 0.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.0 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.7 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |