Project
DANIO-CODE
Navigation
Downloads
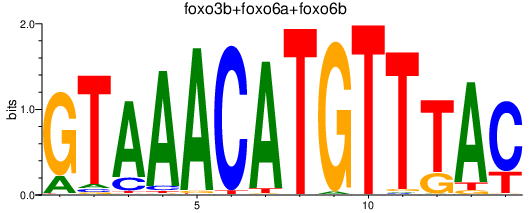
Results for foxo3b+foxo6a+foxo6b
Z-value: 1.48
Transcription factors associated with foxo3b+foxo6a+foxo6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxo6b
|
ENSDARG00000024619 | forkhead box O6 b |
|
foxo3b
|
ENSDARG00000042904 | forkhead box O3b |
|
foxo6a
|
ENSDARG00000100486 | forkhead box O6 a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| foxo3b | dr10_dc_chr20_-_32429820_32429890 | -0.89 | 4.8e-06 | Click! |
| si:ch211-206a7.2 | dr10_dc_chr19_+_15666408_15666441 | -0.78 | 3.8e-04 | Click! |
Activity profile of foxo3b+foxo6a+foxo6b motif
Sorted Z-values of foxo3b+foxo6a+foxo6b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of foxo3b+foxo6a+foxo6b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_19989384 | 5.33 |
ENSDART00000173619
|
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr10_-_25608902 | 4.84 |
ENSDART00000147876
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr15_+_38397772 | 4.53 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr24_-_14446593 | 4.19 |
|
|
|
| chr23_+_28396415 | 3.91 |
ENSDART00000142179
|
birc5b
|
baculoviral IAP repeat containing 5b |
| chr24_+_19270877 | 3.78 |
|
|
|
| chr7_+_29969850 | 3.59 |
ENSDART00000109243
|
sema4bb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Bb |
| chr3_+_44928323 | 3.46 |
ENSDART00000170913
|
zgc:112146
|
zgc:112146 |
| chr7_-_19989419 | 3.41 |
ENSDART00000127699
|
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr5_+_29052293 | 3.36 |
ENSDART00000035400
|
tsc1a
|
tuberous sclerosis 1a |
| chr10_-_7513764 | 3.35 |
ENSDART00000167054
ENSDART00000167706 |
nrg1
|
neuregulin 1 |
| chr5_-_11530803 | 3.31 |
ENSDART00000159896
|
gatsl3
|
GATS protein-like 3 |
| chr5_-_25133229 | 2.83 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr22_-_6916274 | 2.81 |
|
|
|
| chr5_-_17372745 | 2.73 |
ENSDART00000141978
|
si:dkey-112e17.1
|
si:dkey-112e17.1 |
| chr6_+_29763568 | 2.66 |
ENSDART00000151784
|
ptmaa
|
prothymosin, alpha a |
| chr12_+_18849010 | 2.65 |
ENSDART00000078494
|
l3mbtl2
|
l(3)mbt-like 2 (Drosophila) |
| chr9_-_12603178 | 2.55 |
|
|
|
| chr2_-_7260067 | 2.55 |
ENSDART00000092116
|
extl2
|
exostosin-like glycosyltransferase 2 |
| chr12_+_18848952 | 2.51 |
ENSDART00000153235
|
l3mbtl2
|
l(3)mbt-like 2 (Drosophila) |
| chr2_-_37495925 | 2.49 |
ENSDART00000146103
|
dapk3
|
death-associated protein kinase 3 |
| chr7_-_19713757 | 2.43 |
|
|
|
| chr20_+_34091702 | 2.41 |
ENSDART00000061729
|
si:dkey-97o5.1
|
si:dkey-97o5.1 |
| chr14_-_47387481 | 2.32 |
ENSDART00000031498
|
ccna2
|
cyclin A2 |
| chr10_+_33790718 | 2.32 |
ENSDART00000161430
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr21_-_17001108 | 2.32 |
ENSDART00000101263
|
ube2g1b
|
ubiquitin-conjugating enzyme E2G 1b (UBC7 homolog, yeast) |
| chr12_+_47729417 | 2.24 |
ENSDART00000159120
|
polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr7_-_29084775 | 2.15 |
ENSDART00000075757
|
gtf2a2
|
general transcription factor IIA, 2 |
| chr7_+_69211965 | 2.09 |
ENSDART00000028064
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr17_+_28653205 | 2.05 |
ENSDART00000129779
|
hectd1
|
HECT domain containing 1 |
| chr4_-_12782246 | 2.04 |
ENSDART00000058020
|
helb
|
helicase (DNA) B |
| chr8_+_47565879 | 2.04 |
|
|
|
| chr2_+_21698627 | 2.03 |
ENSDART00000133228
|
ctdsplb
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like b |
| chr6_-_22060441 | 2.03 |
ENSDART00000167167
|
utp6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr9_-_12603215 | 2.01 |
|
|
|
| chr15_+_38397715 | 1.97 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr5_+_29052517 | 1.92 |
ENSDART00000035400
|
tsc1a
|
tuberous sclerosis 1a |
| chr7_+_36281268 | 1.91 |
ENSDART00000173682
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr9_+_27909662 | 1.91 |
ENSDART00000112415
|
lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr11_+_24806542 | 1.85 |
ENSDART00000018743
|
phf20a
|
PHD finger protein 20, a |
| chr21_-_41148147 | 1.85 |
ENSDART00000169977
|
bnip1b
|
BCL2/adenovirus E1B interacting protein 1b |
| chr21_-_17001152 | 1.84 |
ENSDART00000101263
|
ube2g1b
|
ubiquitin-conjugating enzyme E2G 1b (UBC7 homolog, yeast) |
| chr13_-_25711537 | 1.83 |
ENSDART00000015154
|
papolg
|
poly(A) polymerase gamma |
| chr16_+_46443800 | 1.82 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr17_-_29005489 | 1.76 |
ENSDART00000157210
|
CR925802.1
|
ENSDARG00000097263 |
| chr5_+_25133592 | 1.76 |
ENSDART00000098467
|
abhd17b
|
abhydrolase domain containing 17B |
| chr7_-_36281088 | 1.74 |
ENSDART00000158224
|
CU464124.1
|
ENSDARG00000103315 |
| chr8_-_53165501 | 1.74 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr25_-_34648876 | 1.73 |
ENSDART00000154851
|
zgc:153405
|
zgc:153405 |
| chr19_-_20026445 | 1.71 |
ENSDART00000160002
|
tax1bp1a
|
Tax1 (human T-cell leukemia virus type I) binding protein 1a |
| chr6_-_41141242 | 1.69 |
ENSDART00000128723
ENSDART00000151055 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr18_+_20571460 | 1.69 |
ENSDART00000151990
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr2_+_22753634 | 1.64 |
ENSDART00000171853
|
LMO4
|
zgc:56628 |
| chr3_+_27163495 | 1.63 |
ENSDART00000175278
ENSDART00000177526 |
BX908803.2
|
ENSDARG00000107836 |
| chr5_+_29052462 | 1.62 |
ENSDART00000035400
|
tsc1a
|
tuberous sclerosis 1a |
| chr2_-_56632969 | 1.62 |
ENSDART00000089158
|
hmha1a
|
histocompatibility (minor) HA-1 a |
| chr7_+_31750808 | 1.56 |
ENSDART00000173848
|
mettl15
|
methyltransferase like 15 |
| chr14_+_46362661 | 1.55 |
ENSDART00000025749
|
ttc9c
|
tetratricopeptide repeat domain 9C |
| chr7_+_29970029 | 1.55 |
ENSDART00000109243
|
sema4bb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Bb |
| chr23_+_28213261 | 1.50 |
ENSDART00000143362
|
BX663515.1
|
ENSDARG00000094837 |
| chr2_-_38097978 | 1.49 |
ENSDART00000056544
|
tox4a
|
TOX high mobility group box family member 4 a |
| chr15_+_38397897 | 1.48 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr16_-_29470341 | 1.46 |
ENSDART00000149289
|
tlr18
|
toll-like receptor 18 |
| chr18_+_20571400 | 1.44 |
ENSDART00000151990
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr6_-_41141280 | 1.42 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr7_+_31750775 | 1.39 |
ENSDART00000173848
|
mettl15
|
methyltransferase like 15 |
| chr23_-_36547458 | 1.38 |
ENSDART00000006881
|
zbtb39
|
zinc finger and BTB domain containing 39 |
| KN150034v1_+_1223 | 1.37 |
|
|
|
| chr3_-_22240424 | 1.36 |
|
|
|
| chr22_+_18294579 | 1.35 |
ENSDART00000033103
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr20_-_7079949 | 1.34 |
ENSDART00000040793
|
sirt5
|
sirtuin 5 |
| chr21_+_17265099 | 1.33 |
ENSDART00000145057
|
tsc1b
|
tuberous sclerosis 1b |
| chr22_+_18294622 | 1.30 |
ENSDART00000033103
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr19_+_20027309 | 1.28 |
ENSDART00000165934
|
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr7_+_69211741 | 1.23 |
ENSDART00000109644
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr12_+_17481598 | 1.22 |
ENSDART00000170449
ENSDART00000111565 |
pms2
|
PMS1 homolog 2, mismatch repair system component |
| chr19_-_7522951 | 1.19 |
ENSDART00000003544
|
gabpb2a
|
GA binding protein transcription factor, beta subunit 2a |
| chr18_+_20571513 | 1.19 |
ENSDART00000136710
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr5_-_11531015 | 1.19 |
ENSDART00000159896
|
gatsl3
|
GATS protein-like 3 |
| chr16_-_29470491 | 1.19 |
ENSDART00000175571
|
tlr18
|
toll-like receptor 18 |
| chr12_-_4444385 | 1.16 |
ENSDART00000092492
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr23_+_28213293 | 1.16 |
ENSDART00000143362
|
BX663515.1
|
ENSDARG00000094837 |
| chr21_-_13026036 | 1.15 |
ENSDART00000135623
|
fam219aa
|
family with sequence similarity 219, member Aa |
| chr19_-_7522853 | 1.14 |
ENSDART00000168194
|
gabpb2a
|
GA binding protein transcription factor, beta subunit 2a |
| chr7_+_69212144 | 1.10 |
ENSDART00000159799
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr13_-_12898069 | 1.09 |
ENSDART00000009499
|
whsc1
|
Wolf-Hirschhorn syndrome candidate 1 |
| chr19_-_13368170 | 1.06 |
ENSDART00000158330
|
zfpm2b
|
zinc finger protein, FOG family member 2b |
| chr24_-_38221052 | 1.05 |
|
|
|
| chr20_-_7079708 | 1.04 |
ENSDART00000040793
|
sirt5
|
sirtuin 5 |
| chr7_-_41601001 | 1.03 |
ENSDART00000174258
|
zgc:92818
|
zgc:92818 |
| chr23_-_903314 | 1.01 |
ENSDART00000165102
|
rhoac
|
ras homolog gene family, member Ac |
| chr20_-_3385325 | 1.00 |
ENSDART00000092264
|
rev3l
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr2_+_22753590 | 0.98 |
ENSDART00000171853
|
LMO4
|
zgc:56628 |
| chr16_-_25485248 | 0.94 |
ENSDART00000040756
|
zgc:136493
|
zgc:136493 |
| chr23_+_44937093 | 0.92 |
|
|
|
| chr12_+_28685081 | 0.91 |
ENSDART00000022724
|
pnpo
|
pyridoxamine 5'-phosphate oxidase |
| KN150034v1_+_1158 | 0.89 |
|
|
|
| chr22_+_2517235 | 0.89 |
ENSDART00000147967
|
si:ch73-92e7.4
|
si:ch73-92e7.4 |
| chr5_-_11530880 | 0.86 |
ENSDART00000159896
|
gatsl3
|
GATS protein-like 3 |
| chr11_+_41307205 | 0.86 |
|
|
|
| chr2_-_38098036 | 0.84 |
ENSDART00000056544
|
tox4a
|
TOX high mobility group box family member 4 a |
| chr24_-_14446522 | 0.84 |
|
|
|
| chr14_-_47387082 | 0.83 |
|
|
|
| chr21_-_19649065 | 0.82 |
ENSDART00000131562
ENSDART00000133467 |
BX927394.1
|
ENSDARG00000092538 |
| chr12_+_47729272 | 0.77 |
ENSDART00000159120
|
polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr6_-_57479549 | 0.76 |
ENSDART00000128065
|
ddx27
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 |
| chr15_-_19398601 | 0.75 |
ENSDART00000062576
|
thyn1
|
thymocyte nuclear protein 1 |
| KN150232v1_+_13079 | 0.74 |
ENSDART00000174701
|
CABZ01084889.1
|
ENSDARG00000107269 |
| chr13_-_10214396 | 0.72 |
ENSDART00000132231
|
AL929457.1
|
ENSDARG00000095483 |
| chr3_+_33788977 | 0.72 |
ENSDART00000055248
|
alkbh7
|
alkB homolog 7 |
| chr2_-_56632913 | 0.70 |
ENSDART00000089158
|
hmha1a
|
histocompatibility (minor) HA-1 a |
| chr8_+_26838596 | 0.70 |
|
|
|
| chr21_-_13026077 | 0.68 |
ENSDART00000024616
|
fam219aa
|
family with sequence similarity 219, member Aa |
| chr5_-_25133456 | 0.64 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr5_-_23171454 | 0.64 |
ENSDART00000135153
|
TBC1D8B
|
TBC1 domain family member 8B |
| chr7_+_36280848 | 0.63 |
ENSDART00000027807
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr7_+_31750514 | 0.62 |
ENSDART00000173976
|
mettl15
|
methyltransferase like 15 |
| chr16_-_29470444 | 0.58 |
ENSDART00000175571
|
tlr18
|
toll-like receptor 18 |
| chr20_+_42993675 | 0.57 |
ENSDART00000153240
ENSDART00000134855 |
efr3bb
|
EFR3 homolog Bb (S. cerevisiae) |
| chr9_+_426701 | 0.56 |
ENSDART00000164310
|
bzw1b
|
basic leucine zipper and W2 domains 1b |
| chr15_-_46501789 | 0.56 |
ENSDART00000099185
|
ilkap
|
integrin-linked kinase-associated serine/threonine phosphatase |
| chr20_+_41904897 | 0.53 |
ENSDART00000139805
|
mcm9
|
minichromosome maintenance 9 homologous recombination repair factor |
| chr11_-_21243422 | 0.53 |
ENSDART00000080116
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr23_-_12223670 | 0.52 |
ENSDART00000135035
|
fam217b
|
family with sequence similarity 217, member B |
| chr25_+_4729095 | 0.51 |
ENSDART00000163839
|
ap4e1
|
adaptor-related protein complex 4, epsilon 1 subunit |
| chr13_-_12898263 | 0.51 |
ENSDART00000009499
|
whsc1
|
Wolf-Hirschhorn syndrome candidate 1 |
| chr22_-_10410949 | 0.51 |
ENSDART00000111962
ENSDART00000064809 |
nol8
|
nucleolar protein 8 |
| chr19_-_20026359 | 0.51 |
ENSDART00000171664
|
tax1bp1a
|
Tax1 (human T-cell leukemia virus type I) binding protein 1a |
| chr13_-_48068159 | 0.50 |
ENSDART00000165157
|
CU468002.1
|
ENSDARG00000103616 |
| chr10_+_45056951 | 0.47 |
ENSDART00000158553
|
ENSDARG00000101143
|
ENSDARG00000101143 |
| chr7_-_19714367 | 0.46 |
ENSDART00000010932
|
snx15
|
sorting nexin 15 |
| chr18_-_21921898 | 0.45 |
ENSDART00000132381
|
edc4
|
enhancer of mRNA decapping 4 |
| chr16_+_29651305 | 0.45 |
ENSDART00000124232
|
mcl1b
|
MCL1, BCL2 family apoptosis regulator b |
| chr15_-_8215797 | 0.43 |
ENSDART00000156663
|
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr3_-_4648706 | 0.42 |
|
|
|
| chr15_-_19398506 | 0.42 |
ENSDART00000062576
|
thyn1
|
thymocyte nuclear protein 1 |
| chr15_-_46501920 | 0.36 |
ENSDART00000099185
|
ilkap
|
integrin-linked kinase-associated serine/threonine phosphatase |
| chr8_-_16428630 | 0.33 |
ENSDART00000098691
|
rnf11b
|
ring finger protein 11b |
| chr5_-_43219303 | 0.32 |
ENSDART00000022481
|
mccc2
|
methylcrotonoyl-CoA carboxylase 2 (beta) |
| chr2_-_44346715 | 0.30 |
ENSDART00000140633
|
sdhc
|
succinate dehydrogenase complex, subunit C, integral membrane protein |
| chr24_-_24884852 | 0.29 |
ENSDART00000122997
|
nup58
|
nucleoporin 58 |
| chr19_+_21783204 | 0.28 |
ENSDART00000024639
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr13_+_48067433 | 0.27 |
ENSDART00000171080
|
msh6
|
mutS homolog 6 (E. coli) |
| chr14_-_6096120 | 0.27 |
|
|
|
| chr15_-_21756778 | 0.26 |
ENSDART00000039865
|
sdhdb
|
succinate dehydrogenase complex, subunit D, integral membrane protein b |
| chr8_+_17975195 | 0.26 |
ENSDART00000039887
ENSDART00000121984 |
ssbp3b
|
single stranded DNA binding protein 3b |
| chr14_-_31675157 | 0.26 |
|
|
|
| chr23_+_17952613 | 0.24 |
ENSDART00000156331
|
si:ch73-390p7.2
|
si:ch73-390p7.2 |
| chr20_+_41904703 | 0.24 |
ENSDART00000139805
|
mcm9
|
minichromosome maintenance 9 homologous recombination repair factor |
| chr16_-_31393328 | 0.23 |
ENSDART00000178298
|
mroh1
|
maestro heat-like repeat family member 1 |
| chr10_-_38582881 | 0.22 |
ENSDART00000157222
|
BX000451.1
|
ENSDARG00000097896 |
| chr18_+_21124045 | 0.22 |
ENSDART00000060191
|
suv420h1
|
suppressor of variegation 4-20 homolog 1 (Drosophila) |
| chr6_-_3837266 | 0.20 |
ENSDART00000059212
|
unc50
|
unc-50 homolog (C. elegans) |
| chr9_+_426850 | 0.19 |
ENSDART00000164310
|
bzw1b
|
basic leucine zipper and W2 domains 1b |
| KN150648v1_+_877 | 0.19 |
|
|
|
| chr25_-_34622553 | 0.18 |
ENSDART00000125128
|
FP236630.1
|
ENSDARG00000086304 |
| chr5_+_38126649 | 0.17 |
ENSDART00000138484
ENSDART00000142867 |
si:dkey-58f10.7
|
si:dkey-58f10.7 |
| chr6_-_57479360 | 0.13 |
ENSDART00000128065
|
ddx27
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 |
| chr3_+_42217445 | 0.13 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr3_+_60367511 | 0.12 |
ENSDART00000128260
|
CABZ01087514.1
|
ENSDARG00000090861 |
| chr6_+_22060547 | 0.12 |
ENSDART00000165271
|
suz12b
|
SUZ12 polycomb repressive complex 2b subunit |
| chr6_+_55835894 | 0.10 |
ENSDART00000108786
|
si:ch211-81n22.1
|
si:ch211-81n22.1 |
| chr18_-_44533391 | 0.09 |
ENSDART00000077125
|
aplp2
|
amyloid beta (A4) precursor-like protein 2 |
| chr16_+_10950751 | 0.06 |
ENSDART00000128762
|
dedd1
|
death effector domain-containing 1 |
| chr15_+_42329433 | 0.06 |
ENSDART00000130404
|
scaf4b
|
SR-related CTD-associated factor 4b |
| chr7_+_25735735 | 0.02 |
ENSDART00000112908
|
pelp1
|
proline, glutamate and leucine rich protein 1 |
| chr23_+_44622486 | 0.01 |
ENSDART00000014691
|
eif4e2rs1
|
eukaryotic translation initiation factor 4E family member 2 related sequence 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0051311 | meiotic metaphase plate congression(GO:0051311) |
| 1.1 | 3.3 | GO:0008347 | glial cell migration(GO:0008347) eurydendroid cell differentiation(GO:0021755) |
| 0.9 | 4.3 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.7 | 8.2 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.6 | 4.4 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.6 | 2.4 | GO:0036048 | regulation of ketone biosynthetic process(GO:0010566) protein demalonylation(GO:0036046) peptidyl-lysine demalonylation(GO:0036047) protein desuccinylation(GO:0036048) peptidyl-lysine desuccinylation(GO:0036049) protein deglutarylation(GO:0061698) |
| 0.6 | 2.3 | GO:0030801 | positive regulation of cyclic nucleotide metabolic process(GO:0030801) positive regulation of cyclic nucleotide biosynthetic process(GO:0030804) positive regulation of nucleotide biosynthetic process(GO:0030810) positive regulation of cyclase activity(GO:0031281) positive regulation of lyase activity(GO:0051349) positive regulation of purine nucleotide biosynthetic process(GO:1900373) |
| 0.5 | 2.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.5 | 1.9 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.3 | 1.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.2 | 0.9 | GO:0042823 | water-soluble vitamin biosynthetic process(GO:0042364) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.2 | 3.6 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.2 | 4.8 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.2 | 3.2 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.1 | 0.6 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 1.8 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 2.1 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 1.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 5.0 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 1.5 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 0.4 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.7 | GO:0070265 | necrotic cell death(GO:0070265) programmed necrotic cell death(GO:0097300) |
| 0.1 | 2.0 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 1.8 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 1.6 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 2.0 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 0.3 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 3.1 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 1.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 2.3 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.4 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 1.7 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 4.2 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.9 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.8 | GO:0032508 | DNA geometric change(GO:0032392) DNA duplex unwinding(GO:0032508) |
| 0.0 | 2.5 | GO:0010942 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.0 | 7.3 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 6.6 | GO:0045892 | negative regulation of transcription, DNA-templated(GO:0045892) |
| 0.0 | 0.1 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 2.6 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.8 | 2.3 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.6 | 4.4 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.4 | 1.2 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.4 | 2.0 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.3 | 2.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 2.6 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.6 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 3.0 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.3 | GO:1905202 | methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.1 | 3.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 1.0 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.3 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.5 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 1.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 2.1 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 1.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.5 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 10.6 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 3.5 | GO:0005730 | nucleolus(GO:0005730) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0071424 | rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) |
| 0.7 | 3.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.7 | 2.7 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.6 | 2.4 | GO:0061697 | protein-malonyllysine demalonylase activity(GO:0036054) protein-succinyllysine desuccinylase activity(GO:0036055) protein-glutaryllysine deglutarylase activity(GO:0061697) |
| 0.5 | 1.8 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.4 | 3.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.4 | 4.8 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.3 | 3.9 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.2 | 0.9 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 2.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.2 | 2.0 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.2 | 5.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.2 | 2.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 1.5 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 1.8 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 5.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 4.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 2.0 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 0.3 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 3.0 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 0.9 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 0.3 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 4.2 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 5.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 2.3 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.1 | 1.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.9 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 2.5 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 3.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.3 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 2.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.7 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 2.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 3.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 3.0 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 2.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 0.5 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 2.1 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.9 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.0 | REACTOME DNA REPAIR | Genes involved in DNA Repair |