Project
DANIO-CODE
Navigation
Downloads
Results for foxo4
Z-value: 1.26
Transcription factors associated with foxo4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxo4
|
ENSDARG00000055792 | forkhead box O4 |
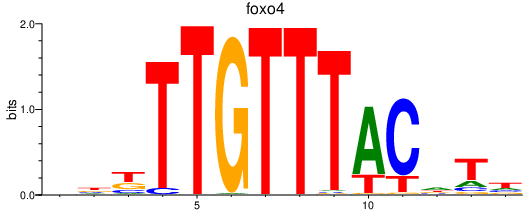
Activity profile of foxo4 motif
Sorted Z-values of foxo4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of foxo4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_32636372 | 5.41 |
ENSDART00000085512
ENSDART00000144694 |
kank1b
|
KN motif and ankyrin repeat domains 1b |
| chr1_-_7160294 | 4.01 |
ENSDART00000174972
|
BX908756.2
|
ENSDARG00000108483 |
| chr15_+_20054306 | 3.88 |
ENSDART00000155199
|
zgc:112083
|
zgc:112083 |
| chr22_-_28828375 | 3.38 |
ENSDART00000104880
ENSDART00000005112 |
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr11_-_25019899 | 3.16 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr1_-_7160099 | 3.13 |
ENSDART00000174972
|
BX908756.2
|
ENSDARG00000108483 |
| chr14_+_14850200 | 3.03 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr8_+_37716781 | 2.95 |
ENSDART00000108556
|
npm2a
|
nucleophosmin/nucleoplasmin, 2a |
| chr16_-_13723352 | 2.83 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr1_-_8593850 | 2.75 |
ENSDART00000146065
ENSDART00000114876 |
ubn1
|
ubinuclein 1 |
| chr13_-_25711537 | 2.73 |
ENSDART00000015154
|
papolg
|
poly(A) polymerase gamma |
| chr7_+_45747622 | 2.43 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr19_+_5399535 | 2.35 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr7_+_45747395 | 2.34 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr9_-_12687806 | 2.31 |
ENSDART00000058565
|
pttg1ipb
|
PTTG1 interacting protein b |
| chr3_-_15325357 | 2.29 |
ENSDART00000139575
|
spns1
|
spinster homolog 1 (Drosophila) |
| chr9_-_21677941 | 2.19 |
ENSDART00000121939
ENSDART00000080404 |
mphosph8
|
M-phase phosphoprotein 8 |
| chr6_-_8008902 | 2.16 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr18_-_20476969 | 2.13 |
ENSDART00000060311
|
paqr5a
|
progestin and adipoQ receptor family member Va |
| chr17_+_14957568 | 2.13 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr25_-_7794186 | 2.11 |
ENSDART00000104686
|
ambra1b
|
autophagy/beclin-1 regulator 1b |
| chr23_-_1562572 | 2.11 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr2_-_30216377 | 2.04 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr16_-_26947117 | 1.99 |
ENSDART00000111156
|
pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr11_-_44859225 | 1.95 |
ENSDART00000163776
|
eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chr17_-_9806363 | 1.93 |
ENSDART00000021942
|
eapp
|
e2f-associated phosphoprotein |
| chr7_+_28824986 | 1.93 |
ENSDART00000052349
|
ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr21_+_20350152 | 1.92 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr16_-_13723295 | 1.92 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr24_+_26223767 | 1.92 |
|
|
|
| chr17_+_33205930 | 1.89 |
ENSDART00000104476
|
snx9a
|
sorting nexin 9a |
| chr23_-_36349563 | 1.89 |
|
|
|
| chr4_-_14208573 | 1.87 |
ENSDART00000015134
|
twf1b
|
twinfilin actin-binding protein 1b |
| chr8_+_26015379 | 1.85 |
ENSDART00000142555
|
arih2
|
ariadne homolog 2 (Drosophila) |
| chr5_+_3567992 | 1.85 |
ENSDART00000129329
|
rpain
|
RPA interacting protein |
| chr17_+_33205732 | 1.82 |
ENSDART00000104476
|
snx9a
|
sorting nexin 9a |
| chr21_+_20350218 | 1.79 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr22_-_29740287 | 1.79 |
ENSDART00000166002
|
pdcd4b
|
programmed cell death 4b |
| chr2_-_30216333 | 1.79 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr11_+_11136919 | 1.78 |
ENSDART00000026135
|
ly75
|
lymphocyte antigen 75 |
| chr5_-_56254482 | 1.75 |
ENSDART00000014028
|
ppm1db
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Db |
| chr13_+_12629211 | 1.74 |
ENSDART00000015127
|
zgc:100846
|
zgc:100846 |
| chr22_-_29740737 | 1.69 |
ENSDART00000009223
|
pdcd4b
|
programmed cell death 4b |
| chr12_-_19063761 | 1.65 |
ENSDART00000153343
|
zc3h7b
|
zinc finger CCCH-type containing 7B |
| chr20_-_53123124 | 1.61 |
ENSDART00000162812
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr7_-_51474531 | 1.61 |
ENSDART00000083190
|
hdac8
|
histone deacetylase 8 |
| chr14_+_41039244 | 1.57 |
ENSDART00000111480
|
bcorl1
|
BCL6 corepressor-like 1 |
| chr5_-_22099244 | 1.55 |
ENSDART00000161298
|
nono
|
non-POU domain containing, octamer-binding |
| chr5_-_66070385 | 1.55 |
ENSDART00000032909
|
kat5b
|
K(lysine) acetyltransferase 5b |
| chr16_-_41764849 | 1.54 |
ENSDART00000084610
|
cep85
|
centrosomal protein 85 |
| chr3_-_26060787 | 1.53 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr3_-_15529158 | 1.53 |
ENSDART00000080441
|
zgc:66443
|
zgc:66443 |
| chr15_+_27000467 | 1.51 |
ENSDART00000023842
|
ppm1da
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Da |
| chr19_-_5186692 | 1.50 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr1_+_50547385 | 1.49 |
ENSDART00000132141
|
btbd3a
|
BTB (POZ) domain containing 3a |
| chr22_+_38322821 | 1.48 |
ENSDART00000104498
|
rcor3
|
REST corepressor 3 |
| chr2_+_58943872 | 1.44 |
ENSDART00000158860
ENSDART00000067736 |
stk11
|
serine/threonine kinase 11 |
| chr23_-_3815871 | 1.44 |
ENSDART00000137826
|
hmga1a
|
high mobility group AT-hook 1a |
| chr15_+_34211736 | 1.42 |
ENSDART00000163513
|
arl4aa
|
ADP-ribosylation factor-like 4aa |
| chr14_+_26417614 | 1.41 |
ENSDART00000148216
|
polr2gl
|
polymerase (RNA) II (DNA directed) polypeptide G-like |
| chr7_+_28825033 | 1.41 |
ENSDART00000052349
|
ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr6_-_8009143 | 1.41 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr22_+_23260371 | 1.40 |
|
|
|
| chr15_+_19948916 | 1.39 |
ENSDART00000054416
|
dyrk1ab
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, b |
| chr15_-_14102102 | 1.37 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr3_+_48810347 | 1.36 |
ENSDART00000133283
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr6_+_38629054 | 1.33 |
ENSDART00000086533
|
atp10a
|
ATPase, class V, type 10A |
| chr21_+_20350334 | 1.33 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr10_-_6587281 | 1.32 |
ENSDART00000163788
ENSDART00000171833 |
chd1
|
chromodomain helicase DNA binding protein 1 |
| chr9_-_37557040 | 1.31 |
ENSDART00000039913
|
hspbap1
|
hspb associated protein 1 |
| chr17_-_23689380 | 1.30 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr10_+_32107014 | 1.30 |
ENSDART00000137373
|
si:ch211-266i6.3
|
si:ch211-266i6.3 |
| chr5_+_44246311 | 1.30 |
ENSDART00000145299
ENSDART00000136521 |
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr25_-_7794262 | 1.29 |
ENSDART00000156761
|
ambra1b
|
autophagy/beclin-1 regulator 1b |
| chr5_-_64432697 | 1.27 |
ENSDART00000165556
|
tor2a
|
torsin family 2, member A |
| chr20_+_53563389 | 1.27 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr16_-_5205600 | 1.26 |
ENSDART00000148955
|
bckdhb
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr24_-_6649190 | 1.25 |
ENSDART00000042478
|
enkur
|
enkurin, TRPC channel interacting protein |
| chr12_+_23691261 | 1.25 |
ENSDART00000066331
|
svila
|
supervillin a |
| chr5_-_56254353 | 1.25 |
ENSDART00000014028
|
ppm1db
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Db |
| chr20_+_1364879 | 1.24 |
ENSDART00000145981
ENSDART00000152709 |
mmachc
|
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria |
| chr22_-_29740663 | 1.24 |
ENSDART00000009223
|
pdcd4b
|
programmed cell death 4b |
| chr8_-_51612689 | 1.23 |
ENSDART00000175779
|
kctd9a
|
potassium channel tetramerization domain containing 9a |
| chr15_-_37687982 | 1.21 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr7_-_51474798 | 1.20 |
ENSDART00000175523
|
hdac8
|
histone deacetylase 8 |
| chr24_-_6649248 | 1.20 |
ENSDART00000042478
|
enkur
|
enkurin, TRPC channel interacting protein |
| chr17_+_32408151 | 1.19 |
ENSDART00000155519
|
si:ch211-139d20.3
|
si:ch211-139d20.3 |
| chr21_+_19511167 | 1.19 |
ENSDART00000058487
|
rai14
|
retinoic acid induced 14 |
| chr18_-_17158316 | 1.16 |
ENSDART00000141873
|
zc3h18
|
zinc finger CCCH-type containing 18 |
| chr22_-_29695242 | 1.16 |
|
|
|
| chr20_-_53123015 | 1.15 |
ENSDART00000162812
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr15_+_19948869 | 1.14 |
ENSDART00000054416
|
dyrk1ab
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, b |
| chr22_+_18840143 | 1.14 |
|
|
|
| chr6_+_40925259 | 1.13 |
ENSDART00000002728
ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr12_+_23691397 | 1.12 |
ENSDART00000066331
|
svila
|
supervillin a |
| chr3_+_32279635 | 1.11 |
ENSDART00000141793
|
rras
|
related RAS viral (r-ras) oncogene homolog |
| chr21_+_34053739 | 1.11 |
ENSDART00000147519
|
mtmr1b
|
myotubularin related protein 1b |
| chr1_+_45148002 | 1.11 |
ENSDART00000148086
|
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr10_-_6587375 | 1.10 |
ENSDART00000163788
ENSDART00000171833 |
chd1
|
chromodomain helicase DNA binding protein 1 |
| chr15_-_31103957 | 1.10 |
ENSDART00000080328
|
nf1a
|
neurofibromin 1a |
| chr20_-_20711298 | 1.09 |
ENSDART00000063492
|
ppm1ab
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Ab |
| chr19_+_40750511 | 1.09 |
ENSDART00000147391
|
fam133b
|
family with sequence similarity 133, member B |
| chr16_-_39317068 | 1.06 |
ENSDART00000133642
|
gpd1l
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr21_-_32048130 | 1.05 |
ENSDART00000137878
|
mat2b
|
methionine adenosyltransferase II, beta |
| chr4_-_1871246 | 1.05 |
ENSDART00000103484
|
gas2l3
|
growth arrest-specific 2 like 3 |
| chr11_+_16017857 | 1.05 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr12_+_19999188 | 1.03 |
|
|
|
| chr11_-_34314863 | 1.02 |
ENSDART00000133302
|
pfkfb4a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4a |
| chr2_-_31728072 | 1.01 |
ENSDART00000113498
|
lrrcc1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr5_-_31474372 | 1.01 |
ENSDART00000162577
|
arpc5lb
|
actin related protein 2/3 complex, subunit 5-like, b |
| chr11_-_25019298 | 1.01 |
ENSDART00000130477
ENSDART00000079578 |
snai1a
|
snail family zinc finger 1a |
| chr8_+_26015350 | 1.00 |
ENSDART00000004521
|
arih2
|
ariadne homolog 2 (Drosophila) |
| chr23_+_35551931 | 1.00 |
ENSDART00000132609
|
ccnt1
|
cyclin T1 |
| chr15_-_28654137 | 1.00 |
ENSDART00000156049
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr20_+_26005068 | 0.99 |
ENSDART00000016864
|
slc35f6
|
solute carrier family 35, member F6 |
| chr18_+_38963423 | 0.99 |
|
|
|
| chr12_+_18794467 | 0.99 |
ENSDART00000127536
|
cbx7b
|
chromobox homolog 7b |
| chr23_+_38313746 | 0.99 |
ENSDART00000129593
|
znf217
|
zinc finger protein 217 |
| chr19_+_10477101 | 0.98 |
ENSDART00000151735
|
necap1
|
NECAP endocytosis associated 1 |
| chr23_+_39718769 | 0.98 |
ENSDART00000034690
|
otud3
|
OTU deubiquitinase 3 |
| chr23_+_22052173 | 0.98 |
ENSDART00000087110
|
eif4g3b
|
eukaryotic translation initiation factor 4 gamma, 3b |
| chr17_+_43916865 | 0.98 |
ENSDART00000049503
|
peli2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr17_-_9806413 | 0.98 |
ENSDART00000021942
|
eapp
|
e2f-associated phosphoprotein |
| chr13_-_33096839 | 0.97 |
ENSDART00000057386
|
golga5
|
golgin A5 |
| chr22_+_23260413 | 0.97 |
|
|
|
| chr24_+_14095601 | 0.97 |
ENSDART00000124740
|
ncoa2
|
nuclear receptor coactivator 2 |
| chr20_-_21906340 | 0.97 |
ENSDART00000145807
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr5_-_18393705 | 0.94 |
ENSDART00000010101
ENSDART00000099434 |
aacs
|
acetoacetyl-CoA synthetase |
| chr3_-_15529108 | 0.92 |
ENSDART00000080441
|
zgc:66443
|
zgc:66443 |
| chr2_-_45118469 | 0.92 |
ENSDART00000018818
|
mul1a
|
mitochondrial E3 ubiquitin protein ligase 1a |
| chr23_+_32015527 | 0.92 |
ENSDART00000088607
|
nemp1
|
nuclear envelope integral membrane protein 1 |
| chr15_-_37687790 | 0.92 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr8_-_25696708 | 0.91 |
ENSDART00000007482
|
tspy
|
testis specific protein, Y-linked |
| chr3_+_19536018 | 0.90 |
ENSDART00000006490
|
tlk2
|
tousled-like kinase 2 |
| chr19_-_33013672 | 0.90 |
ENSDART00000134149
|
zgc:91944
|
zgc:91944 |
| chr3_+_52745606 | 0.88 |
ENSDART00000104683
|
pbx4
|
pre-B-cell leukemia transcription factor 4 |
| chr13_+_28487504 | 0.88 |
ENSDART00000087001
|
cnnm2a
|
cyclin and CBS domain divalent metal cation transport mediator 2a |
| chr15_+_45738963 | 0.88 |
|
|
|
| chr22_-_28828300 | 0.88 |
ENSDART00000104880
|
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr5_-_56254422 | 0.87 |
ENSDART00000014028
|
ppm1db
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Db |
| chr6_-_8009055 | 0.86 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr13_+_969851 | 0.86 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| chr4_+_8669695 | 0.83 |
ENSDART00000168768
|
adipor2
|
adiponectin receptor 2 |
| chr15_+_25745737 | 0.83 |
ENSDART00000077853
|
hic1
|
hypermethylated in cancer 1 |
| chr14_+_2820269 | 0.82 |
ENSDART00000162445
|
hmgxb3
|
HMG box domain containing 3 |
| chr1_+_8594006 | 0.82 |
ENSDART00000142384
|
BX323038.1
|
ENSDARG00000092791 |
| chr12_-_28422582 | 0.81 |
ENSDART00000067762
|
MYO1D
|
myosin ID |
| chr15_+_27000663 | 0.81 |
ENSDART00000023842
|
ppm1da
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Da |
| chr4_-_4247251 | 0.81 |
ENSDART00000150279
|
cd9b
|
CD9 molecule b |
| chr19_-_10737772 | 0.80 |
ENSDART00000081379
|
olah
|
oleoyl-ACP hydrolase |
| chr23_+_22052045 | 0.80 |
ENSDART00000145172
|
eif4g3b
|
eukaryotic translation initiation factor 4 gamma, 3b |
| chr13_-_22772707 | 0.80 |
ENSDART00000089133
|
rufy2
|
RUN and FYVE domain containing 2 |
| chr15_-_37687938 | 0.79 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr21_+_34053590 | 0.79 |
ENSDART00000147519
ENSDART00000158115 ENSDART00000029599 ENSDART00000145123 |
mtmr1b
|
myotubularin related protein 1b |
| chr17_+_33205888 | 0.79 |
ENSDART00000104476
|
snx9a
|
sorting nexin 9a |
| chr23_+_44470297 | 0.79 |
ENSDART00000149842
|
MEPCE
|
methylphosphate capping enzyme |
| chr21_-_9689702 | 0.79 |
ENSDART00000158836
|
arhgap24
|
Rho GTPase activating protein 24 |
| chr3_-_26060702 | 0.78 |
ENSDART00000144726
|
ypel3
|
yippee-like 3 |
| chr17_+_10592507 | 0.78 |
ENSDART00000097274
|
ATG14
|
autophagy related 14 |
| chr5_+_44538762 | 0.77 |
ENSDART00000172702
ENSDART00000136002 |
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr10_-_6587666 | 0.77 |
ENSDART00000158881
|
chd1
|
chromodomain helicase DNA binding protein 1 |
| chr20_-_16949608 | 0.77 |
ENSDART00000027582
|
brms1lb
|
breast cancer metastasis-suppressor 1-like b |
| chr5_+_36904127 | 0.77 |
ENSDART00000165465
|
si:ch1073-224n8.1
|
si:ch1073-224n8.1 |
| chr7_-_24557143 | 0.77 |
ENSDART00000138193
|
otub1b
|
OTU deubiquitinase, ubiquitin aldehyde binding 1b |
| chr13_-_22772577 | 0.77 |
ENSDART00000089133
|
rufy2
|
RUN and FYVE domain containing 2 |
| chr3_-_26060748 | 0.75 |
ENSDART00000144726
|
ypel3
|
yippee-like 3 |
| chr3_+_10182994 | 0.75 |
ENSDART00000156700
|
cbx2
|
chromobox homolog 2 (Drosophila Pc class) |
| chr2_-_30216455 | 0.75 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr7_+_72030256 | 0.75 |
ENSDART00000172021
|
tollip
|
toll interacting protein |
| chr2_+_33206305 | 0.74 |
ENSDART00000145588
|
rnf220a
|
ring finger protein 220a |
| chr23_-_36350288 | 0.74 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
| chr3_-_15325178 | 0.74 |
ENSDART00000139575
|
spns1
|
spinster homolog 1 (Drosophila) |
| chr3_+_10182687 | 0.74 |
ENSDART00000156700
|
cbx2
|
chromobox homolog 2 (Drosophila Pc class) |
| chr3_+_54777037 | 0.73 |
ENSDART00000132587
|
mpg
|
N-methylpurine DNA glycosylase |
| chr17_+_43729816 | 0.72 |
ENSDART00000164097
|
ENSDARG00000101112
|
ENSDARG00000101112 |
| chr15_+_27000248 | 0.72 |
ENSDART00000023842
|
ppm1da
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Da |
| chr19_-_25842574 | 0.71 |
ENSDART00000036854
|
glcci1
|
glucocorticoid induced 1 |
| KN149858v1_+_13014 | 0.71 |
ENSDART00000165916
|
MED8
|
mediator complex subunit 8 |
| chr16_-_55263338 | 0.70 |
ENSDART00000113358
|
wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr1_+_50547341 | 0.70 |
ENSDART00000132141
|
btbd3a
|
BTB (POZ) domain containing 3a |
| chr8_+_12913443 | 0.70 |
ENSDART00000081601
|
cept1a
|
choline/ethanolamine phosphotransferase 1a |
| chr12_+_43683149 | 0.69 |
ENSDART00000162014
|
si:ch211-232h12.2
|
si:ch211-232h12.2 |
| chr22_+_37696341 | 0.68 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr10_-_25755035 | 0.67 |
ENSDART00000140483
|
scaf4a
|
SR-related CTD-associated factor 4a |
| chr2_-_30216703 | 0.67 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr2_+_17027403 | 0.67 |
ENSDART00000164329
|
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr10_-_39272645 | 0.67 |
ENSDART00000161758
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr8_-_25696852 | 0.67 |
ENSDART00000007482
|
tspy
|
testis specific protein, Y-linked |
| chr9_+_24398747 | 0.66 |
ENSDART00000090280
ENSDART00000133479 |
zgc:153901
|
zgc:153901 |
| chr10_+_39272734 | 0.66 |
ENSDART00000075943
|
ei24
|
etoposide induced 2.4 |
| chr24_-_26224123 | 0.66 |
ENSDART00000080039
|
lrrc31
|
leucine rich repeat containing 31 |
| chr20_+_53563189 | 0.66 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr24_-_5903682 | 0.66 |
ENSDART00000007373
ENSDART00000135124 |
acbd5a
|
acyl-CoA binding domain containing 5a |
| chr10_+_573116 | 0.66 |
ENSDART00000129856
ENSDART00000110384 |
smad4a
|
SMAD family member 4a |
| chr13_-_42274444 | 0.65 |
ENSDART00000043069
|
march5
|
membrane-associated ring finger (C3HC4) 5 |
| chr2_+_11422195 | 0.65 |
ENSDART00000019078
|
lhx8a
|
LIM homeobox 8a |
| chr2_-_24413154 | 0.64 |
ENSDART00000145526
|
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr14_+_8288174 | 0.64 |
ENSDART00000037749
|
stx5a
|
syntaxin 5A |
| chr4_+_5147577 | 0.64 |
ENSDART00000067392
|
tigarb
|
tp53-induced glycolysis and apoptosis regulator b |
| chr20_-_3258772 | 0.63 |
ENSDART00000166831
|
rps6kc1
|
ribosomal protein S6 kinase polypeptide 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.3 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.5 | 1.5 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.4 | 2.8 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.4 | 2.8 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.4 | 2.2 | GO:0036268 | swimming(GO:0036268) |
| 0.3 | 1.7 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.3 | 0.9 | GO:0032024 | positive regulation of peptide secretion(GO:0002793) positive regulation of insulin secretion(GO:0032024) positive regulation of hormone secretion(GO:0046887) positive regulation of peptide hormone secretion(GO:0090277) |
| 0.3 | 1.9 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.2 | 2.0 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.2 | 1.0 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.2 | 1.3 | GO:0050428 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 | 1.1 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.2 | 0.6 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) hepatic duct development(GO:0061011) |
| 0.2 | 2.1 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.2 | 3.0 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.2 | 1.4 | GO:0014009 | glial cell proliferation(GO:0014009) regulation of glial cell proliferation(GO:0060251) |
| 0.2 | 0.7 | GO:0051055 | negative regulation of lipid biosynthetic process(GO:0051055) |
| 0.2 | 4.7 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.2 | 1.1 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.2 | 0.7 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 4.7 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.2 | 1.0 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.2 | 0.6 | GO:1900543 | negative regulation of nucleotide metabolic process(GO:0045980) negative regulation of purine nucleotide metabolic process(GO:1900543) |
| 0.2 | 0.8 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.2 | 7.8 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.1 | 0.6 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.8 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 1.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.5 | GO:0018008 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.1 | 1.5 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.8 | GO:1902914 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 2.7 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 0.6 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 1.2 | GO:0035794 | positive regulation of mitochondrial membrane permeability(GO:0035794) mitochondrial outer membrane permeabilization(GO:0097345) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.1 | 0.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 1.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.7 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.3 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 1.2 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 | 4.7 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.1 | 1.1 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 0.4 | GO:0035313 | negative regulation of Rac protein signal transduction(GO:0035021) wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 4.6 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 1.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.3 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 0.8 | GO:0007584 | response to nutrient(GO:0007584) |
| 0.1 | 0.4 | GO:0000423 | macromitophagy(GO:0000423) |
| 0.1 | 2.9 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 4.0 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 0.5 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 2.0 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 3.2 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.1 | 1.0 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 2.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.3 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 1.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.9 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.6 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.6 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 1.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.8 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.9 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.0 | 1.9 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 1.0 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 2.0 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.2 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.0 | 0.3 | GO:0071850 | positive regulation of cytokinesis(GO:0032467) mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.4 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 2.1 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 1.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.5 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 1.9 | GO:0071599 | otic vesicle development(GO:0071599) |
| 0.0 | 0.7 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.2 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.5 | GO:0071910 | determination of pancreatic left/right asymmetry(GO:0035469) determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.7 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.1 | GO:0003342 | bulbus arteriosus development(GO:0003232) proepicardium development(GO:0003342) septum transversum development(GO:0003343) |
| 0.0 | 0.1 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.0 | 1.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.1 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.4 | GO:0051307 | meiotic chromosome separation(GO:0051307) |
| 0.0 | 0.8 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 2.1 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.4 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.4 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.6 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.6 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 1.8 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 3.0 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.8 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.4 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.7 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.8 | GO:0048916 | posterior lateral line development(GO:0048916) |
| 0.0 | 0.1 | GO:0007032 | endosome organization(GO:0007032) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.8 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.4 | 2.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.3 | 1.4 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.3 | 0.8 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 2.0 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.2 | 1.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 0.8 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 2.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.3 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 0.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.8 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.0 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 1.5 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 6.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 1.0 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 1.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.7 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.0 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 2.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.1 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.3 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 1.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.8 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 1.0 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 3.6 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.6 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 5.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 13.0 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 0.1 | GO:0000784 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.5 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.5 | GO:0031201 | SNARE complex(GO:0031201) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.9 | 2.8 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.7 | 2.7 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.5 | 1.5 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.3 | 6.7 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.3 | 0.8 | GO:0048270 | methionine adenosyltransferase regulator activity(GO:0048270) |
| 0.2 | 2.8 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.2 | 0.7 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.2 | 1.3 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.2 | 1.9 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 1.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.2 | 1.2 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.2 | 1.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.2 | 3.2 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.2 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 0.6 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.2 | 1.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.9 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 2.0 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 1.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 0.5 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.1 | 1.0 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 3.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.8 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.1 | 2.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 2.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 1.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 4.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 2.3 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.1 | 1.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.6 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.7 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.1 | 0.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.4 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 1.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.9 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.9 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 1.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 2.1 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 2.0 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 1.1 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 5.1 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 2.0 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 2.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 5.4 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.8 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 2.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 5.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.9 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.7 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 2.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 2.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.3 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.4 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 1.0 | GO:0042803 | protein homodimerization activity(GO:0042803) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 1.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 1.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 0.9 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 1.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 0.7 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 3.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 4.8 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.2 | 1.7 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.2 | 1.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 1.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 2.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.0 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 0.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 1.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.0 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.5 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 0.9 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 1.9 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 2.1 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.4 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.0 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 8.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.8 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |