Project
DANIO-CODE
Navigation
Downloads
Results for foxq2
Z-value: 0.47
Transcription factors associated with foxq2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
foxq2
|
ENSDARG00000071394 | forkhead box Q2 |
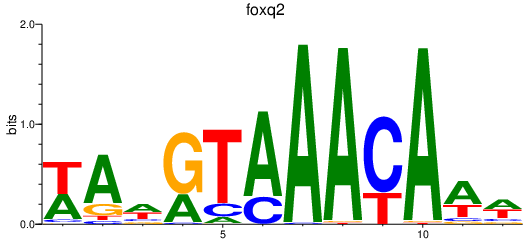
Activity profile of foxq2 motif
Sorted Z-values of foxq2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of foxq2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_32838287 | 0.73 |
|
|
|
| chr15_-_23711689 | 0.68 |
ENSDART00000128644
|
ckmb
|
creatine kinase, muscle b |
| chr6_-_14821305 | 0.52 |
|
|
|
| chr22_-_29740737 | 0.47 |
ENSDART00000009223
|
pdcd4b
|
programmed cell death 4b |
| chr23_-_30861356 | 0.45 |
ENSDART00000114628
|
myt1a
|
myelin transcription factor 1a |
| chr2_+_30932612 | 0.37 |
ENSDART00000132450
ENSDART00000137012 |
myom1a
|
myomesin 1a (skelemin) |
| chr14_+_38375351 | 0.37 |
ENSDART00000175744
|
BX088707.3
|
ENSDARG00000107351 |
| chr21_+_10775209 | 0.32 |
|
|
|
| chr4_-_1871246 | 0.32 |
ENSDART00000103484
|
gas2l3
|
growth arrest-specific 2 like 3 |
| chr11_+_30416115 | 0.32 |
ENSDART00000161662
|
ttbk1a
|
tau tubulin kinase 1a |
| chr5_+_49093134 | 0.32 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr11_+_6809190 | 0.31 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr12_+_17744208 | 0.31 |
|
|
|
| chr13_+_28723589 | 0.30 |
|
|
|
| chr23_-_1562603 | 0.29 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr25_-_18855316 | 0.29 |
ENSDART00000155927
|
si:ch211-68a17.7
|
si:ch211-68a17.7 |
| chr8_-_24092176 | 0.28 |
ENSDART00000099692
|
dclre1b
|
DNA cross-link repair 1B |
| chr15_+_28269289 | 0.28 |
ENSDART00000077736
|
vtna
|
vitronectin a |
| chr7_+_24787515 | 0.28 |
|
|
|
| chr23_+_10535195 | 0.28 |
ENSDART00000140557
|
tns2a
|
tensin 2a |
| chr16_-_26947117 | 0.28 |
ENSDART00000111156
|
pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr21_-_27845076 | 0.27 |
ENSDART00000132583
|
nrxn2a
|
neurexin 2a |
| chr6_-_9329917 | 0.27 |
ENSDART00000151470
|
map3k2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr18_-_16806665 | 0.26 |
ENSDART00000048722
|
ampd3b
|
adenosine monophosphate deaminase 3b |
| chr6_+_4712785 | 0.26 |
ENSDART00000151674
|
pcdh9
|
protocadherin 9 |
| chr13_-_42274486 | 0.26 |
ENSDART00000043069
|
march5
|
membrane-associated ring finger (C3HC4) 5 |
| chr23_+_10534968 | 0.25 |
ENSDART00000140557
|
tns2a
|
tensin 2a |
| chr12_+_22282872 | 0.25 |
ENSDART00000153055
|
hdlbpb
|
high density lipoprotein binding protein b |
| chr23_-_1562572 | 0.25 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr2_+_25904910 | 0.25 |
ENSDART00000133623
|
pld1a
|
phospholipase D1a |
| chr5_+_49093250 | 0.25 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr17_-_23689233 | 0.25 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr8_-_13183034 | 0.24 |
ENSDART00000090541
|
si:ch73-61d6.3
|
si:ch73-61d6.3 |
| chr4_-_18660286 | 0.24 |
ENSDART00000178638
|
pparaa
|
peroxisome proliferator-activated receptor alpha a |
| chr20_-_10133201 | 0.24 |
ENSDART00000144970
|
meis2b
|
Meis homeobox 2b |
| chr4_+_10722966 | 0.24 |
ENSDART00000102534
ENSDART00000067253 ENSDART00000136000 |
stab2
|
stabilin 2 |
| chr25_+_18855775 | 0.23 |
ENSDART00000123207
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr25_+_18855868 | 0.23 |
ENSDART00000154291
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr18_-_6896957 | 0.23 |
ENSDART00000175747
|
ppp6r2b
|
protein phosphatase 6, regulatory subunit 2b |
| chr6_+_24717985 | 0.23 |
ENSDART00000165609
|
barhl2
|
BarH-like homeobox 2 |
| chr9_-_45104857 | 0.23 |
ENSDART00000131252
|
zgc:66484
|
zgc:66484 |
| chr13_-_17938316 | 0.22 |
ENSDART00000146772
ENSDART00000134477 |
zfand4
|
zinc finger, AN1-type domain 4 |
| chr24_+_30343717 | 0.22 |
ENSDART00000162377
|
FP085414.1
|
ENSDARG00000100270 |
| chr6_-_6895376 | 0.22 |
ENSDART00000081761
|
bin1b
|
bridging integrator 1b |
| chr5_-_25509188 | 0.22 |
ENSDART00000137376
|
BX510923.1
|
ENSDARG00000094488 |
| chr20_-_39371023 | 0.22 |
ENSDART00000127173
|
clu
|
clusterin |
| chr16_-_2972770 | 0.21 |
ENSDART00000082522
|
abcd2
|
ATP-binding cassette, sub-family D (ALD), member 2 |
| chr2_+_38178934 | 0.21 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr19_-_33013672 | 0.21 |
ENSDART00000134149
|
zgc:91944
|
zgc:91944 |
| chr2_+_27674700 | 0.21 |
ENSDART00000087643
|
tesk2
|
testis-specific kinase 2 |
| chr24_+_19065679 | 0.20 |
ENSDART00000139299
|
zgc:162928
|
zgc:162928 |
| chr1_-_44199728 | 0.20 |
ENSDART00000145354
|
tcirg1a
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3a |
| chr24_-_19573615 | 0.20 |
ENSDART00000158952
ENSDART00000109107 ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr14_+_151136 | 0.20 |
ENSDART00000165766
|
mcm7
|
minichromosome maintenance complex component 7 |
| chr7_+_9726412 | 0.20 |
ENSDART00000173155
|
adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr17_-_23689317 | 0.20 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr21_+_11592175 | 0.19 |
ENSDART00000147473
|
pcsk1
|
proprotein convertase subtilisin/kexin type 1 |
| chr15_+_34211736 | 0.19 |
ENSDART00000163513
|
arl4aa
|
ADP-ribosylation factor-like 4aa |
| chr14_-_33704021 | 0.19 |
ENSDART00000149396
ENSDART00000123607 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr3_-_42985563 | 0.19 |
ENSDART00000176655
|
CU138533.3
|
ENSDARG00000108986 |
| chr6_-_40660116 | 0.19 |
ENSDART00000154359
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr5_-_18990190 | 0.18 |
ENSDART00000098795
|
mmab
|
methylmalonic aciduria (cobalamin deficiency) cblB type |
| chr3_-_56803756 | 0.18 |
ENSDART00000014103
|
zgc:112148
|
zgc:112148 |
| chr6_-_31160132 | 0.18 |
ENSDART00000153592
|
CR847887.1
|
ENSDARG00000097226 |
| chr14_-_15777250 | 0.18 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr19_-_33013748 | 0.18 |
ENSDART00000134149
|
zgc:91944
|
zgc:91944 |
| chr7_+_39116005 | 0.18 |
ENSDART00000146702
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr14_-_29459799 | 0.18 |
ENSDART00000005568
|
pdlim3b
|
PDZ and LIM domain 3b |
| chr5_+_7691621 | 0.18 |
ENSDART00000161261
|
lmbrd2a
|
LMBR1 domain containing 2a |
| chr9_+_33343936 | 0.18 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr5_-_28693064 | 0.17 |
|
|
|
| chr5_-_25509259 | 0.17 |
ENSDART00000137376
|
BX510923.1
|
ENSDARG00000094488 |
| chr3_-_33773025 | 0.16 |
|
|
|
| chr1_+_31210417 | 0.16 |
ENSDART00000007522
|
anos1a
|
anosmin 1a |
| chr9_-_6683651 | 0.16 |
ENSDART00000061593
|
pou3f3a
|
POU class 3 homeobox 3a |
| chr2_+_5651428 | 0.16 |
ENSDART00000111220
|
mb21d2
|
Mab-21 domain containing 2 |
| chr2_-_30216455 | 0.16 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr5_+_15880920 | 0.15 |
ENSDART00000168643
|
CABZ01088700.1
|
ENSDARG00000101452 |
| chr5_+_37053530 | 0.15 |
ENSDART00000161051
|
sptbn2
|
spectrin, beta, non-erythrocytic 2 |
| chr18_+_19430116 | 0.15 |
ENSDART00000025107
|
map2k1
|
mitogen-activated protein kinase kinase 1 |
| chr10_+_10770717 | 0.15 |
ENSDART00000146910
|
swi5
|
SWI5 homologous recombination repair protein |
| chr7_-_66641971 | 0.15 |
|
|
|
| chr25_-_13990944 | 0.15 |
ENSDART00000124140
|
zgc:101566
|
zgc:101566 |
| chr3_-_28534446 | 0.15 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr1_+_53294559 | 0.15 |
ENSDART00000159900
|
ccsapa
|
centriole, cilia and spindle-associated protein a |
| chr22_+_18294622 | 0.15 |
ENSDART00000033103
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr16_+_14697969 | 0.14 |
ENSDART00000133566
|
deptor
|
DEP domain containing MTOR-interacting protein |
| chr23_+_24557956 | 0.14 |
|
|
|
| chr22_-_29740663 | 0.14 |
ENSDART00000009223
|
pdcd4b
|
programmed cell death 4b |
| chr8_-_17132057 | 0.14 |
ENSDART00000143920
|
mrps36
|
mitochondrial ribosomal protein S36 |
| chr24_+_30343687 | 0.14 |
ENSDART00000162377
|
FP085414.1
|
ENSDARG00000100270 |
| chr16_-_1736470 | 0.14 |
ENSDART00000124660
|
ascc3
|
activating signal cointegrator 1 complex subunit 3 |
| chr25_-_34677352 | 0.14 |
ENSDART00000115210
|
ano9a
|
anoctamin 9a |
| chr25_+_35159867 | 0.14 |
|
|
|
| chr25_+_33924376 | 0.14 |
|
|
|
| chr14_-_15776937 | 0.14 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr13_-_30515486 | 0.14 |
ENSDART00000139607
|
zcchc24
|
zinc finger, CCHC domain containing 24 |
| chr24_+_37752868 | 0.14 |
ENSDART00000158181
|
wdr24
|
WD repeat domain 24 |
| chr2_-_30216377 | 0.13 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr13_-_17729510 | 0.13 |
ENSDART00000170583
|
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr10_+_39148251 | 0.13 |
ENSDART00000125986
|
ENSDARG00000055339
|
ENSDARG00000055339 |
| chr17_+_24595050 | 0.13 |
ENSDART00000064738
|
atpif1b
|
ATPase inhibitory factor 1b |
| chr2_-_30216333 | 0.13 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr13_+_30546398 | 0.13 |
|
|
|
| chr13_-_42274444 | 0.13 |
ENSDART00000043069
|
march5
|
membrane-associated ring finger (C3HC4) 5 |
| chr7_-_50827308 | 0.13 |
ENSDART00000121574
|
col4a6
|
collagen, type IV, alpha 6 |
| chr18_+_26916897 | 0.13 |
ENSDART00000050230
|
tspan3a
|
tetraspanin 3a |
| chr7_+_24787720 | 0.12 |
|
|
|
| chr15_+_25745737 | 0.12 |
ENSDART00000077853
|
hic1
|
hypermethylated in cancer 1 |
| chr14_+_16632329 | 0.12 |
ENSDART00000163013
|
limch1b
|
LIM and calponin homology domains 1b |
| chr15_+_23615727 | 0.12 |
ENSDART00000152720
|
MARK4
|
microtubule affinity regulating kinase 4 |
| chr23_-_18851683 | 0.12 |
ENSDART00000169180
ENSDART00000006482 |
trpc4apb
|
transient receptor potential cation channel, subfamily C, member 4 associated protein b |
| chr6_-_6895175 | 0.12 |
ENSDART00000149232
ENSDART00000150033 |
bin1b
|
bridging integrator 1b |
| chr7_-_8465929 | 0.11 |
ENSDART00000172928
|
tex261
|
testis expressed 261 |
| chr25_-_14953284 | 0.11 |
ENSDART00000165632
|
pax6a
|
paired box 6a |
| chr4_-_14927871 | 0.11 |
ENSDART00000110199
|
prdm4
|
PR domain containing 4 |
| chr8_-_2557556 | 0.11 |
ENSDART00000140033
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr20_-_23272102 | 0.11 |
ENSDART00000167197
|
spata18
|
spermatogenesis associated 18 |
| chr9_-_19154264 | 0.11 |
ENSDART00000081878
|
pou1f1
|
POU class 1 homeobox 1 |
| chr13_+_1377875 | 0.11 |
|
|
|
| KN150520v1_+_33937 | 0.10 |
|
|
|
| chr18_-_9980662 | 0.10 |
|
|
|
| chr15_+_3136812 | 0.10 |
ENSDART00000130968
|
rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr8_-_13182911 | 0.10 |
ENSDART00000090541
|
si:ch73-61d6.3
|
si:ch73-61d6.3 |
| chr5_-_11723101 | 0.10 |
ENSDART00000161706
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr21_+_8334453 | 0.10 |
ENSDART00000055336
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr10_+_516885 | 0.10 |
ENSDART00000128275
|
npffr1l3
|
neuropeptide FF receptor 1 like 3 |
| chr22_-_29740287 | 0.10 |
ENSDART00000166002
|
pdcd4b
|
programmed cell death 4b |
| chr11_-_26463925 | 0.10 |
ENSDART00000083010
|
acad9
|
acyl-CoA dehydrogenase family, member 9 |
| chr18_-_6897024 | 0.10 |
ENSDART00000135206
|
ppp6r2b
|
protein phosphatase 6, regulatory subunit 2b |
| chr8_+_49582361 | 0.09 |
ENSDART00000108613
|
rasef
|
RAS and EF-hand domain containing |
| chr22_-_5624894 | 0.09 |
ENSDART00000165221
|
mcm2
|
minichromosome maintenance complex component 2 |
| chr11_-_38856849 | 0.09 |
ENSDART00000113185
|
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr1_+_41148042 | 0.09 |
ENSDART00000145170
ENSDART00000136879 |
smox
|
spermine oxidase |
| chr24_-_25316192 | 0.09 |
ENSDART00000105820
|
mbtps2
|
membrane-bound transcription factor peptidase, site 2 |
| chr1_+_35919158 | 0.09 |
ENSDART00000176692
|
CR762429.1
|
ENSDARG00000108193 |
| chr3_+_48810347 | 0.09 |
ENSDART00000133283
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr3_+_26015378 | 0.09 |
|
|
|
| chr19_-_24551474 | 0.09 |
ENSDART00000104087
|
thap7
|
THAP domain containing 7 |
| chr2_+_1645259 | 0.09 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr15_+_22510499 | 0.09 |
ENSDART00000040542
|
arhgef12a
|
Rho guanine nucleotide exchange factor (GEF) 12a |
| chr19_-_13045465 | 0.09 |
ENSDART00000128975
|
slc25a32a
|
solute carrier family 25 (mitochondrial folate carrier), member 32a |
| chr2_-_30216703 | 0.09 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr23_-_36350288 | 0.09 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
| chr17_-_42778156 | 0.09 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr24_+_9163779 | 0.09 |
|
|
|
| chr7_+_9659464 | 0.09 |
ENSDART00000172824
|
cers3a
|
ceramide synthase 3a |
| chr25_+_13566205 | 0.09 |
ENSDART00000008989
|
ccdc113
|
coiled-coil domain containing 113 |
| chr14_-_17270741 | 0.08 |
ENSDART00000161355
ENSDART00000168959 |
rnf4
|
ring finger protein 4 |
| chr1_-_53465275 | 0.08 |
|
|
|
| chr14_-_2430754 | 0.08 |
ENSDART00000159004
|
si:ch73-233f7.8
|
si:ch73-233f7.8 |
| chr21_+_5775383 | 0.08 |
ENSDART00000165065
|
uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr21_+_38685219 | 0.08 |
ENSDART00000076157
|
rab24
|
RAB24, member RAS oncogene family |
| chr22_-_11490627 | 0.08 |
ENSDART00000063157
|
slc26a11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr3_+_40113121 | 0.08 |
ENSDART00000074746
|
smcr8a
|
Smith-Magenis syndrome chromosome region, candidate 8a |
| chr14_+_14850200 | 0.08 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr19_+_7630847 | 0.07 |
ENSDART00000151758
|
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr16_+_48674191 | 0.07 |
ENSDART00000004751
|
pbx2
|
pre-B-cell leukemia homeobox 2 |
| chr15_-_30935719 | 0.07 |
ENSDART00000050649
|
msi2b
|
musashi RNA-binding protein 2b |
| chr7_-_53947785 | 0.07 |
ENSDART00000165875
|
csnk1g1
|
casein kinase 1, gamma 1 |
| chr11_-_13283709 | 0.07 |
ENSDART00000164978
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr1_-_50395003 | 0.07 |
ENSDART00000035150
|
spast
|
spastin |
| chr2_+_27674825 | 0.07 |
ENSDART00000087643
|
tesk2
|
testis-specific kinase 2 |
| chr22_+_18840143 | 0.07 |
|
|
|
| chr21_+_27379622 | 0.07 |
ENSDART00000108763
|
cfb
|
complement factor B |
| chr4_-_17735989 | 0.07 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr25_-_16890343 | 0.07 |
ENSDART00000073419
|
ccnd2a
|
cyclin D2, a |
| chr23_-_27681694 | 0.07 |
ENSDART00000026314
|
phf8
|
PHD finger protein 8 |
| chr7_-_8465988 | 0.07 |
ENSDART00000172928
|
tex261
|
testis expressed 261 |
| chr1_-_7160099 | 0.07 |
ENSDART00000174972
|
BX908756.2
|
ENSDARG00000108483 |
| chr16_-_41181493 | 0.07 |
ENSDART00000102649
ENSDART00000145956 |
ptpn23a
|
protein tyrosine phosphatase, non-receptor type 23, a |
| chr8_+_23464087 | 0.07 |
ENSDART00000026316
|
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr18_-_48375325 | 0.06 |
|
|
|
| chr23_+_13777809 | 0.06 |
ENSDART00000144386
|
uckl1b
|
uridine-cytidine kinase 1-like 1b |
| chr13_-_46423391 | 0.06 |
|
|
|
| chr5_-_62592627 | 0.06 |
ENSDART00000131274
|
tbcelb
|
tubulin folding cofactor E-like b |
| chr3_-_2010512 | 0.06 |
|
|
|
| chr25_-_13774670 | 0.06 |
ENSDART00000160866
|
cry2
|
cryptochrome circadian clock 2 |
| chr18_+_35861883 | 0.06 |
ENSDART00000131121
|
ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr18_+_19430014 | 0.06 |
ENSDART00000025107
|
map2k1
|
mitogen-activated protein kinase kinase 1 |
| chr20_+_52740555 | 0.06 |
ENSDART00000110777
|
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr10_+_39148457 | 0.06 |
ENSDART00000125986
|
ENSDARG00000055339
|
ENSDARG00000055339 |
| chr14_-_1081316 | 0.06 |
|
|
|
| chr18_+_19430260 | 0.06 |
ENSDART00000025107
|
map2k1
|
mitogen-activated protein kinase kinase 1 |
| chr7_+_13386796 | 0.06 |
ENSDART00000157870
|
clpxa
|
caseinolytic mitochondrial matrix peptidase chaperone subunit a |
| chr22_-_14103803 | 0.06 |
ENSDART00000062902
|
ENSDARG00000042857
|
ENSDARG00000042857 |
| chr3_+_5665300 | 0.05 |
ENSDART00000101807
|
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr23_+_31073628 | 0.05 |
ENSDART00000085263
|
SELENOI
|
selenoprotein I |
| chr17_-_23689380 | 0.05 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr4_-_20414881 | 0.05 |
ENSDART00000161343
|
cep83
|
centrosomal protein 83 |
| chr10_-_32936408 | 0.05 |
ENSDART00000138243
|
rabgef1
|
RAB guanine nucleotide exchange factor (GEF) 1 |
| chr8_-_20830316 | 0.05 |
ENSDART00000147267
|
si:ch211-133l5.8
|
si:ch211-133l5.8 |
| chr11_+_41324391 | 0.05 |
ENSDART00000173103
|
aldh4a1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr14_+_21401757 | 0.05 |
ENSDART00000073707
|
stx3a
|
syntaxin 3A |
| chr16_-_43432131 | 0.05 |
ENSDART00000111963
|
atad2
|
ATPase family, AAA domain containing 2 |
| chr4_+_70917419 | 0.05 |
ENSDART00000174035
|
si:cabz01054394.5
|
si:cabz01054394.5 |
| chr25_-_14953370 | 0.05 |
ENSDART00000159490
|
pax6a
|
paired box 6a |
| chr10_+_4987461 | 0.05 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr7_-_8466131 | 0.05 |
ENSDART00000065488
|
tex261
|
testis expressed 261 |
| chr3_+_54777037 | 0.04 |
ENSDART00000132587
|
mpg
|
N-methylpurine DNA glycosylase |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.5 | GO:0050428 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.3 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.1 | 0.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.1 | 0.8 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.1 | 0.2 | GO:1901492 | regulation of lymphangiogenesis(GO:1901490) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.3 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.1 | 0.2 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.1 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.5 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.3 | GO:0032530 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.1 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.2 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) |
| 0.0 | 0.3 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.2 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.3 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.2 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.2 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.7 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.0 | 0.3 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.2 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.2 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.1 | GO:0043243 | positive regulation of protein complex disassembly(GO:0043243) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0007045 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 0.2 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) |
| 0.0 | 0.0 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.3 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.1 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.0 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.3 | GO:0005844 | polysome(GO:0005844) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 0.7 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.5 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.2 | GO:0097020 | COPII adaptor activity(GO:0097020) |
| 0.1 | 0.5 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 0.3 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 0.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 0.4 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.2 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.1 | GO:0032034 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.1 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.3 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0015245 | long-chain fatty acid transporter activity(GO:0005324) fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.2 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |