Project
DANIO-CODE
Navigation
Downloads
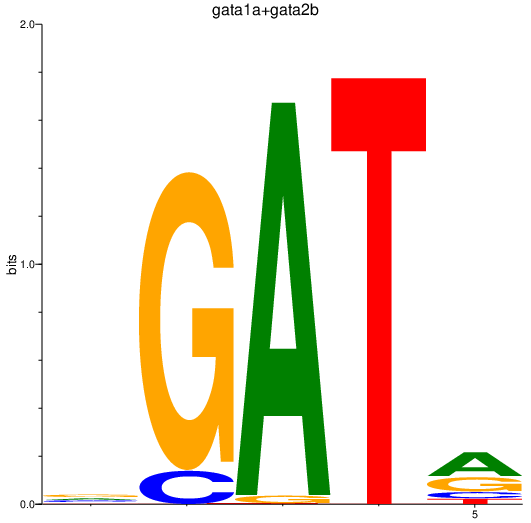
Results for gata1a+gata2b
Z-value: 0.21
Transcription factors associated with gata1a+gata2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gata2b
|
ENSDARG00000009094 | GATA binding protein 2b |
|
gata1a
|
ENSDARG00000013477 | GATA binding protein 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gata1a | dr10_dc_chr11_-_25181234_25181253 | -0.65 | 6.1e-03 | Click! |
Activity profile of gata1a+gata2b motif
Sorted Z-values of gata1a+gata2b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of gata1a+gata2b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_21373501 | 0.47 |
ENSDART00000089408
|
shdb
|
Src homology 2 domain containing transforming protein D, b |
| chr6_-_49160475 | 0.42 |
ENSDART00000041942
|
tspan2a
|
tetraspanin 2a |
| chr22_-_11817641 | 0.34 |
ENSDART00000000192
|
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr17_+_23534953 | 0.33 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr5_-_6054581 | 0.28 |
|
|
|
| chr6_+_9735100 | 0.23 |
ENSDART00000171518
|
abi2b
|
abl-interactor 2b |
| chr22_-_16244177 | 0.22 |
ENSDART00000105681
|
cdc14ab
|
cell division cycle 14Ab |
| chr8_-_6782167 | 0.20 |
ENSDART00000135834
ENSDART00000172157 |
dock5
|
dedicator of cytokinesis 5 |
| chr17_+_23534824 | 0.18 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr4_-_1496779 | 0.15 |
ENSDART00000166360
|
nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr5_+_17543117 | 0.12 |
ENSDART00000132164
|
hira
|
histone cell cycle regulator a |
| chr19_+_12525407 | 0.11 |
ENSDART00000135706
|
ldlrad4a
|
low density lipoprotein receptor class A domain containing 4a |
| chr25_+_32085705 | 0.11 |
ENSDART00000132698
|
ctdspl2a
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2a |
| chr6_-_49160398 | 0.08 |
ENSDART00000143241
|
tspan2a
|
tetraspanin 2a |
| chr10_+_42761028 | 0.05 |
ENSDART00000075259
|
fgfr1b
|
fibroblast growth factor receptor 1b |
| chr20_+_34126950 | 0.01 |
ENSDART00000134305
|
prg4b
|
proteoglycan 4b |
| chr7_+_15081529 | 0.01 |
ENSDART00000006018
|
mespaa
|
mesoderm posterior aa |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.1 | GO:0060394 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0070412 | R-SMAD binding(GO:0070412) |