Project
DANIO-CODE
Navigation
Downloads
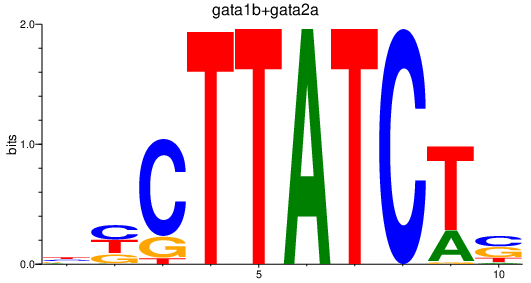
Results for gata1b+gata2a
Z-value: 3.21
Transcription factors associated with gata1b+gata2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gata1b
|
ENSDARG00000059130 | GATA binding protein 1b |
|
gata2a
|
ENSDARG00000059327 | GATA binding protein 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gata2a | dr10_dc_chr11_-_3846064_3846121 | 0.88 | 8.0e-06 | Click! |
Activity profile of gata1b+gata2a motif
Sorted Z-values of gata1b+gata2a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of gata1b+gata2a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_25157675 | 12.48 |
ENSDART00000136984
|
fabp2
|
fatty acid binding protein 2, intestinal |
| chr23_+_7445760 | 10.92 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr7_-_50827308 | 10.68 |
ENSDART00000121574
|
col4a6
|
collagen, type IV, alpha 6 |
| chr20_+_15653121 | 10.22 |
ENSDART00000152702
|
jun
|
jun proto-oncogene |
| chr25_+_21732255 | 10.14 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr15_+_19902697 | 9.77 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr10_+_10843384 | 8.78 |
ENSDART00000130283
|
ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr13_+_2774422 | 8.54 |
ENSDART00000162362
|
wu:fj16a03
|
wu:fj16a03 |
| chr1_+_29054033 | 8.13 |
ENSDART00000054066
|
zic2b
|
zic family member 2 (odd-paired homolog, Drosophila) b |
| chr11_-_28367271 | 7.21 |
ENSDART00000065853
|
dhrs3b
|
dehydrogenase/reductase (SDR family) member 3b |
| chr2_+_4366169 | 7.20 |
ENSDART00000157903
|
gata6
|
GATA binding protein 6 |
| chr23_+_36023748 | 7.17 |
|
|
|
| chr12_-_5628401 | 6.72 |
ENSDART00000105884
|
itga3b
|
integrin, alpha 3b |
| chr10_+_13251463 | 6.67 |
ENSDART00000000887
|
rassf6
|
Ras association (RalGDS/AF-6) domain family 6 |
| chr6_+_58921655 | 6.57 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr8_-_50299273 | 6.55 |
ENSDART00000023639
|
nkx2.7
|
NK2 transcription factor related 7 |
| chr7_+_34018862 | 6.46 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr23_+_25782195 | 6.37 |
ENSDART00000060059
|
rbms2b
|
RNA binding motif, single stranded interacting protein 2b |
| chr23_-_10747605 | 6.23 |
|
|
|
| chr8_-_40287789 | 6.21 |
ENSDART00000074125
|
aplnra
|
apelin receptor a |
| chr23_-_24755654 | 6.21 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr16_-_30606364 | 5.92 |
ENSDART00000147986
|
lmna
|
lamin A |
| chr23_-_23474703 | 5.54 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr21_-_22510751 | 5.49 |
ENSDART00000169870
|
myo5b
|
myosin VB |
| chr11_-_1374026 | 5.48 |
ENSDART00000172953
|
rpl29
|
ribosomal protein L29 |
| chr25_+_18467217 | 5.36 |
ENSDART00000170841
|
cav1
|
caveolin 1 |
| chr8_+_7740132 | 5.30 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr4_-_24298444 | 5.25 |
ENSDART00000077926
ENSDART00000128368 |
celf2
|
cugbp, Elav-like family member 2 |
| chr10_-_17527124 | 5.22 |
ENSDART00000137905
|
nt5c2l1
|
5'-nucleotidase, cytosolic II, like 1 |
| chr12_-_20228689 | 5.10 |
ENSDART00000066384
|
hbbe2
|
hemoglobin beta embryonic-2 |
| chr25_+_7858886 | 4.92 |
ENSDART00000171904
|
ucmab
|
upper zone of growth plate and cartilage matrix associated b |
| chr8_-_18502159 | 4.78 |
ENSDART00000148802
ENSDART00000149081 ENSDART00000148962 |
nexn
|
nexilin (F actin binding protein) |
| chr22_-_36915567 | 4.76 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr22_+_16509286 | 4.63 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr5_+_34022151 | 4.63 |
ENSDART00000141338
|
enc1
|
ectodermal-neural cortex 1 |
| chr20_-_22478716 | 4.58 |
ENSDART00000110967
ENSDART00000011135 |
kita
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog a |
| chr19_+_4996328 | 4.54 |
ENSDART00000165082
|
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr7_-_73530653 | 4.53 |
ENSDART00000166633
ENSDART00000009888 ENSDART00000171254 |
casq1b
|
calsequestrin 1b |
| chr13_+_22350043 | 4.53 |
ENSDART00000136863
|
ldb3a
|
LIM domain binding 3a |
| chr16_+_55351732 | 4.50 |
ENSDART00000109391
|
CABZ01052290.1
|
ENSDARG00000077559 |
| chr6_-_39278328 | 4.40 |
ENSDART00000148531
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr16_+_51316772 | 4.39 |
ENSDART00000169022
|
hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr8_+_15968469 | 4.39 |
ENSDART00000134787
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr3_-_56969897 | 4.34 |
|
|
|
| chr3_+_24145201 | 4.30 |
ENSDART00000169765
|
pdgfbb
|
platelet-derived growth factor beta polypeptide b |
| chr3_+_28371725 | 4.07 |
ENSDART00000151081
|
sept12
|
septin 12 |
| chr12_+_34026544 | 4.00 |
ENSDART00000153413
|
cyth1b
|
cytohesin 1b |
| chr8_+_23464087 | 3.97 |
ENSDART00000026316
|
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| KN149726v1_+_1094 | 3.97 |
|
|
|
| chr18_+_27753985 | 3.96 |
ENSDART00000141835
|
tspan18b
|
tetraspanin 18b |
| chr11_-_28367400 | 3.92 |
ENSDART00000065853
|
dhrs3b
|
dehydrogenase/reductase (SDR family) member 3b |
| chr25_-_13631707 | 3.84 |
ENSDART00000169865
|
lcat
|
lecithin-cholesterol acyltransferase |
| chr13_-_22901327 | 3.82 |
ENSDART00000056523
|
hkdc1
|
hexokinase domain containing 1 |
| chr1_-_16876855 | 3.80 |
ENSDART00000125125
|
acsl1a
|
acyl-CoA synthetase long-chain family member 1a |
| chr8_-_40425193 | 3.79 |
ENSDART00000130798
|
myl7
|
myosin, light chain 7, regulatory |
| chr8_+_48488009 | 3.76 |
|
|
|
| chr19_-_6856033 | 3.75 |
ENSDART00000170952
|
pvrl2l
|
poliovirus receptor-related 2 like |
| chr20_+_40247918 | 3.70 |
ENSDART00000121818
|
trdn
|
triadin |
| chr11_-_13069266 | 3.68 |
ENSDART00000169052
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr3_+_52901602 | 3.64 |
ENSDART00000114343
|
brd4
|
bromodomain containing 4 |
| chr6_+_54231519 | 3.61 |
ENSDART00000149542
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr17_-_42108276 | 3.59 |
ENSDART00000110871
ENSDART00000155484 |
nkx2.4a
|
NK2 homeobox 4a |
| chr17_-_14663139 | 3.53 |
ENSDART00000037371
|
ppp1r13ba
|
protein phosphatase 1, regulatory subunit 13Ba |
| chr5_-_63487161 | 3.52 |
ENSDART00000048395
|
cmlc1
|
cardiac myosin light chain-1 |
| chr10_-_43200499 | 3.45 |
ENSDART00000171494
|
ssbp2
|
single-stranded DNA binding protein 2 |
| chr17_-_13072982 | 3.39 |
ENSDART00000178859
|
CABZ01047998.1
|
ENSDARG00000105945 |
| chr11_-_1792616 | 3.39 |
|
|
|
| chr16_-_13652342 | 3.39 |
ENSDART00000144140
|
BX323458.2
|
ENSDARG00000094614 |
| chr9_+_33167554 | 3.38 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr20_+_33972327 | 3.33 |
ENSDART00000170930
|
rxrgb
|
retinoid X receptor, gamma b |
| chr9_+_49054601 | 3.30 |
ENSDART00000047401
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr25_-_35698109 | 3.26 |
ENSDART00000134928
|
nfatc3b
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3b |
| chr11_-_37921642 | 3.22 |
|
|
|
| chr21_-_40607549 | 3.19 |
ENSDART00000172706
|
pcyt1bb
|
phosphate cytidylyltransferase 1, choline, beta b |
| chr25_+_18467835 | 3.15 |
ENSDART00000172338
|
cav1
|
caveolin 1 |
| chr16_+_13534390 | 3.09 |
ENSDART00000157396
|
ENSDARG00000022807
|
ENSDARG00000022807 |
| chr16_+_55121654 | 3.08 |
ENSDART00000149449
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr4_+_1680973 | 3.07 |
ENSDART00000149826
ENSDART00000166437 ENSDART00000031135 ENSDART00000172300 ENSDART00000067446 |
slc38a4
|
solute carrier family 38, member 4 |
| chr16_+_26576242 | 3.05 |
ENSDART00000039746
|
epb41b
|
erythrocyte membrane protein band 4.1b |
| chr24_-_11768200 | 3.04 |
ENSDART00000033621
|
tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr14_+_26247858 | 3.02 |
ENSDART00000105932
|
si:dkeyp-110e4.11
|
si:dkeyp-110e4.11 |
| chr5_+_14703190 | 2.99 |
ENSDART00000040826
|
tbx1
|
T-box 1 |
| chr24_-_39970858 | 2.99 |
ENSDART00000134251
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
| chr1_+_25662910 | 2.95 |
ENSDART00000113020
|
tet2
|
tet methylcytosine dioxygenase 2 |
| chr6_-_43679553 | 2.94 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr5_+_37996820 | 2.93 |
ENSDART00000015136
|
mogat3b
|
monoacylglycerol O-acyltransferase 3b |
| chr6_-_586251 | 2.93 |
ENSDART00000148867
ENSDART00000149414 ENSDART00000149248 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr11_+_31022048 | 2.93 |
ENSDART00000160154
|
si:dkey-238i5.2
|
si:dkey-238i5.2 |
| chr16_-_31835463 | 2.91 |
ENSDART00000148389
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr21_-_9313788 | 2.83 |
|
|
|
| chr8_-_3974357 | 2.82 |
ENSDART00000163754
ENSDART00000169474 |
mtmr3
|
myotubularin related protein 3 |
| chr18_+_27753913 | 2.74 |
ENSDART00000141835
|
tspan18b
|
tetraspanin 18b |
| chr18_+_48433986 | 2.72 |
ENSDART00000076861
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr8_+_6533379 | 2.66 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr8_-_25678416 | 2.64 |
ENSDART00000143938
|
sema3ga
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ga |
| chr24_+_19374099 | 2.62 |
ENSDART00000056081
ENSDART00000027022 |
sulf1
|
sulfatase 1 |
| chr25_-_3730472 | 2.60 |
|
|
|
| chr17_-_26849495 | 2.59 |
ENSDART00000153590
|
si:dkey-221l4.10
|
si:dkey-221l4.10 |
| chr16_-_13652300 | 2.57 |
ENSDART00000144140
|
BX323458.2
|
ENSDARG00000094614 |
| chr15_-_18110169 | 2.46 |
|
|
|
| chr2_-_52202004 | 2.40 |
ENSDART00000165350
|
BX908782.1
|
ENSDARG00000098957 |
| chr20_+_25567046 | 2.37 |
ENSDART00000153071
|
pfas
|
phosphoribosylformylglycinamidine synthase |
| chr10_+_13251650 | 2.37 |
ENSDART00000136932
|
rassf6
|
Ras association (RalGDS/AF-6) domain family 6 |
| chr23_+_36016450 | 2.37 |
ENSDART00000103035
|
hoxc6a
|
homeobox C6a |
| chr14_+_28126921 | 2.30 |
ENSDART00000026846
|
pin4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| KN150171v1_+_3909 | 2.29 |
ENSDART00000171048
|
CABZ01071639.1
|
ENSDARG00000099435 |
| chr24_+_28441089 | 2.27 |
|
|
|
| chr8_+_48888672 | 2.26 |
|
|
|
| chr11_-_25181234 | 2.25 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr14_-_27754549 | 2.23 |
ENSDART00000135337
|
zgc:64189
|
zgc:64189 |
| chr24_-_25546068 | 2.22 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr10_+_10780578 | 2.22 |
ENSDART00000170147
ENSDART00000004181 |
slc27a4
|
solute carrier family 27 (fatty acid transporter), member 4 |
| chr16_-_22860052 | 2.21 |
ENSDART00000127570
|
pbxip1b
|
pre-B-cell leukemia homeobox interacting protein 1b |
| chr7_-_15004167 | 2.21 |
ENSDART00000030009
|
ENSDARG00000023868
|
ENSDARG00000023868 |
| chr8_+_29733109 | 2.20 |
ENSDART00000020621
|
mapk4
|
mitogen-activated protein kinase 4 |
| chr25_+_18467034 | 2.20 |
ENSDART00000170841
|
cav1
|
caveolin 1 |
| chr22_-_36915720 | 2.19 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr14_+_50011198 | 2.19 |
ENSDART00000171955
|
CR855307.1
|
ENSDARG00000099628 |
| chr21_+_39918070 | 2.17 |
ENSDART00000135235
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr23_+_25930072 | 2.16 |
ENSDART00000124103
|
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr16_+_55121164 | 2.16 |
ENSDART00000149264
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr2_+_4366568 | 2.14 |
ENSDART00000166476
|
gata6
|
GATA binding protein 6 |
| chr2_+_38279372 | 2.10 |
ENSDART00000076478
|
dhrs1
|
dehydrogenase/reductase (SDR family) member 1 |
| chr4_+_7499654 | 2.10 |
ENSDART00000158999
|
tnnt2e
|
troponin T2e, cardiac |
| chr3_-_35472632 | 2.09 |
ENSDART00000055269
|
gng13b
|
guanine nucleotide binding protein (G protein), gamma 13b |
| chr7_+_44106327 | 2.05 |
ENSDART00000108766
ENSDART00000111441 |
cdh5
|
cadherin 5 |
| chr10_+_21693418 | 2.05 |
ENSDART00000160754
|
pcdh1g1
|
protocadherin 1 gamma 1 |
| chr3_-_26073676 | 2.04 |
ENSDART00000169123
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr5_+_52198613 | 2.01 |
ENSDART00000162459
|
scarb2a
|
scavenger receptor class B, member 2a |
| chr12_+_34615991 | 1.96 |
|
|
|
| chr21_-_11741819 | 1.96 |
ENSDART00000081652
|
rfesd
|
Rieske (Fe-S) domain containing |
| chr22_-_11596329 | 1.93 |
ENSDART00000063133
|
gcga
|
glucagon a |
| chr13_+_24132494 | 1.92 |
ENSDART00000135992
|
abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr24_-_25545773 | 1.91 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr8_+_20592039 | 1.90 |
ENSDART00000160421
|
nfic
|
nuclear factor I/C |
| chr22_+_21095535 | 1.89 |
|
|
|
| chr8_-_14113377 | 1.88 |
ENSDART00000090427
|
si:dkey-6n6.1
|
si:dkey-6n6.1 |
| chr3_-_26073923 | 1.87 |
ENSDART00000103748
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr12_+_33879261 | 1.80 |
|
|
|
| chr10_-_36603263 | 1.79 |
ENSDART00000108484
ENSDART00000086861 |
nipblb
|
nipped-B homolog b (Drosophila) |
| chr11_-_35313757 | 1.78 |
ENSDART00000031441
|
sema3fb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fb |
| chr16_+_31890795 | 1.74 |
ENSDART00000159821
|
atn1
|
atrophin 1 |
| chr22_-_17026421 | 1.73 |
ENSDART00000138382
|
nfia
|
nuclear factor I/A |
| chr7_+_9659464 | 1.72 |
ENSDART00000172824
|
cers3a
|
ceramide synthase 3a |
| chr5_-_68024401 | 1.70 |
ENSDART00000016307
|
her1
|
hairy-related 1 |
| chr2_+_4366515 | 1.66 |
ENSDART00000166476
|
gata6
|
GATA binding protein 6 |
| chr16_-_24753560 | 1.65 |
ENSDART00000063083
|
pon3.2
|
paraoxonase 3, tandem duplicate 2 |
| chr12_-_13042845 | 1.64 |
ENSDART00000152510
|
ccser2b
|
coiled-coil serine-rich protein 2b |
| chr1_+_25974174 | 1.63 |
ENSDART00000152144
|
si:dkey-25o16.2
|
si:dkey-25o16.2 |
| chr3_-_12087518 | 1.63 |
ENSDART00000081374
|
cfap70
|
cilia and flagella associated protein 70 |
| chr7_+_26980284 | 1.54 |
ENSDART00000173962
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr13_+_45339261 | 1.52 |
ENSDART00000045329
|
mgst3b
|
microsomal glutathione S-transferase 3b |
| chr5_+_26127909 | 1.52 |
ENSDART00000098545
|
tmem150aa
|
transmembrane protein 150Aa |
| chr22_-_17026124 | 1.51 |
ENSDART00000138382
|
nfia
|
nuclear factor I/A |
| chr1_+_11485480 | 1.45 |
ENSDART00000132560
|
stra6l
|
STRA6-like |
| chr18_-_16806665 | 1.44 |
ENSDART00000048722
|
ampd3b
|
adenosine monophosphate deaminase 3b |
| chr11_+_43136945 | 1.43 |
|
|
|
| chr25_-_21697789 | 1.41 |
|
|
|
| chr5_-_12025080 | 1.40 |
ENSDART00000164190
|
ksr2
|
kinase suppressor of ras 2 |
| chr2_-_30787161 | 1.33 |
ENSDART00000131230
|
rgs20
|
regulator of G protein signaling 20 |
| chr8_+_22559432 | 1.32 |
|
|
|
| chr5_+_36295353 | 1.31 |
ENSDART00000135776
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr8_-_43568078 | 1.29 |
ENSDART00000051139
ENSDART00000160289 |
ncor2
|
nuclear receptor corepressor 2 |
| chr14_-_32063302 | 1.29 |
ENSDART00000173114
|
fgf13a
|
fibroblast growth factor 13a |
| chr1_+_25974337 | 1.28 |
ENSDART00000152617
|
si:dkey-25o16.2
|
si:dkey-25o16.2 |
| chr20_+_19093073 | 1.27 |
ENSDART00000147105
|
tdh
|
L-threonine dehydrogenase |
| chr14_-_31918174 | 1.22 |
ENSDART00000052949
|
fgf13a
|
fibroblast growth factor 13a |
| chr10_+_19568414 | 1.21 |
ENSDART00000162912
|
vsig8a
|
V-set and immunoglobulin domain containing 8a |
| chr23_-_17524325 | 1.18 |
ENSDART00000104680
|
tpd52l2b
|
tumor protein D52-like 2b |
| chr10_-_175160 | 1.17 |
|
|
|
| chr1_-_23659549 | 1.14 |
ENSDART00000177989
|
lrba
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr25_+_12542563 | 1.13 |
|
|
|
| chr8_+_15968636 | 1.12 |
ENSDART00000141173
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr18_+_48434068 | 1.12 |
ENSDART00000076861
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr10_-_22988906 | 1.11 |
|
|
|
| chr20_+_40248154 | 1.11 |
ENSDART00000121818
|
trdn
|
triadin |
| chr1_+_25974264 | 1.09 |
ENSDART00000152785
|
si:dkey-25o16.2
|
si:dkey-25o16.2 |
| chr5_-_13145749 | 1.01 |
ENSDART00000166957
|
purba
|
purine-rich element binding protein Ba |
| chr22_-_22696091 | 1.00 |
|
|
|
| chr2_+_38279454 | 0.98 |
ENSDART00000076478
|
dhrs1
|
dehydrogenase/reductase (SDR family) member 1 |
| chr19_-_46058123 | 0.91 |
|
|
|
| chr16_+_14820248 | 0.90 |
|
|
|
| chr21_-_25719352 | 0.90 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr3_-_29932835 | 0.90 |
|
|
|
| chr24_+_35173816 | 0.89 |
|
|
|
| chr24_-_17089437 | 0.88 |
|
|
|
| chr19_+_38270817 | 0.86 |
|
|
|
| chr22_-_36915689 | 0.84 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr15_-_19162052 | 0.82 |
|
|
|
| chr8_-_25678085 | 0.81 |
ENSDART00000143938
|
sema3ga
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ga |
| chr13_+_24132236 | 0.81 |
ENSDART00000088005
|
abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr6_-_58921392 | 0.79 |
|
|
|
| chr19_+_39100820 | 0.76 |
|
|
|
| chr13_-_25643419 | 0.69 |
ENSDART00000145640
|
pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr16_+_31890895 | 0.69 |
ENSDART00000159821
|
atn1
|
atrophin 1 |
| chr18_+_27753844 | 0.67 |
ENSDART00000141835
|
tspan18b
|
tetraspanin 18b |
| chr15_+_31956131 | 0.67 |
|
|
|
| chr5_-_24247047 | 0.66 |
|
|
|
| chr25_-_22955066 | 0.60 |
ENSDART00000024633
|
dusp8a
|
dual specificity phosphatase 8a |
| chr7_-_65012626 | 0.58 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.9 | GO:0048618 | post-embryonic morphogenesis(GO:0009886) post-embryonic foregut morphogenesis(GO:0048618) |
| 2.7 | 10.7 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 2.2 | 6.6 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 1.8 | 5.5 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 1.5 | 4.6 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 1.5 | 10.7 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) dendritic cell differentiation(GO:0097028) |
| 1.5 | 4.5 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 1.4 | 9.8 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 1.3 | 10.1 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 1.3 | 3.8 | GO:0055004 | atrial cardiac myofibril assembly(GO:0055004) |
| 1.2 | 4.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 1.1 | 7.8 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 1.1 | 11.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 1.1 | 4.3 | GO:1903392 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 1.0 | 8.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 1.0 | 3.0 | GO:0061323 | cell proliferation involved in heart morphogenesis(GO:0061323) regulation of cell proliferation involved in heart morphogenesis(GO:2000136) |
| 0.9 | 3.6 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.9 | 7.2 | GO:0014831 | gastro-intestinal system smooth muscle contraction(GO:0014831) |
| 0.9 | 5.3 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.8 | 5.9 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.8 | 6.5 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.8 | 4.1 | GO:0051281 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) positive regulation of calcium ion transport into cytosol(GO:0010524) positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904323) positive regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904325) |
| 0.8 | 9.0 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.7 | 2.2 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.7 | 8.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.7 | 2.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.6 | 10.7 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.6 | 3.0 | GO:0006211 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 0.6 | 3.5 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.6 | 2.2 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.5 | 3.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.5 | 3.6 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.5 | 5.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.5 | 2.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.4 | 4.9 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.4 | 2.2 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.3 | 3.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.3 | 2.1 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.3 | 2.6 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.3 | 1.5 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.3 | 1.3 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.3 | 3.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 | 7.5 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.2 | 2.0 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 1.4 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.2 | 2.8 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.2 | 4.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 5.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.2 | 3.3 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.2 | 2.4 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.2 | 2.2 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.2 | 9.2 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.2 | 1.4 | GO:0006188 | IMP biosynthetic process(GO:0006188) IMP salvage(GO:0032264) |
| 0.2 | 10.0 | GO:0033334 | fin morphogenesis(GO:0033334) |
| 0.1 | 5.5 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.1 | 1.9 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.5 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.1 | 12.5 | GO:0033993 | response to lipid(GO:0033993) |
| 0.1 | 2.9 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 0.4 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 3.1 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.6 | GO:0034405 | response to fluid shear stress(GO:0034405) |
| 0.1 | 1.7 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 3.6 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 1.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 1.2 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.1 | 7.4 | GO:0045765 | regulation of angiogenesis(GO:0045765) |
| 0.1 | 2.1 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.1 | 4.2 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.1 | 5.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 2.6 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 4.5 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 5.0 | GO:0034101 | erythrocyte differentiation(GO:0030218) erythrocyte homeostasis(GO:0034101) |
| 0.0 | 0.9 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 2.9 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 1.7 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 5.2 | GO:0006417 | regulation of translation(GO:0006417) |
| 0.0 | 3.2 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 5.2 | GO:0006260 | DNA replication(GO:0006260) |
| 0.0 | 1.9 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 0.2 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 4.2 | GO:0050877 | neurological system process(GO:0050877) |
| 0.0 | 9.0 | GO:0042981 | regulation of apoptotic process(GO:0042981) |
| 0.0 | 6.8 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 2.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 2.9 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 1.0 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 2.3 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.4 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.7 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.9 | 4.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.9 | 2.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.7 | 5.1 | GO:0005833 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.5 | 4.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.3 | 11.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.3 | 10.7 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.2 | 4.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 6.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.2 | 8.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.2 | 4.1 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.2 | 5.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 5.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.6 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.1 | 5.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 4.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 2.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 14.5 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 2.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 9.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 2.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 14.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 5.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 52.4 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 3.6 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 20.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 4.0 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 3.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 3.6 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 3.1 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 3.5 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 2.3 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 4.4 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 1.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 10.5 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 8.2 | GO:1990904 | ribonucleoprotein complex(GO:1990904) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 11.1 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 1.3 | 10.1 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 1.1 | 12.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 1.1 | 11.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.8 | 3.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.8 | 3.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.7 | 2.9 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.7 | 5.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.7 | 4.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.7 | 3.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.7 | 5.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.6 | 3.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.5 | 4.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.5 | 2.6 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.5 | 6.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.4 | 7.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.4 | 19.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.4 | 1.3 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.4 | 4.1 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.4 | 1.7 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.4 | 3.8 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.4 | 2.9 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.4 | 2.2 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.3 | 2.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.3 | 3.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.3 | 5.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.3 | 1.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 4.8 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.3 | 2.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.3 | 2.1 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.3 | 2.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 2.8 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.3 | 1.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 5.2 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.2 | 2.4 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.2 | 2.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 7.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.2 | 9.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 10.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 8.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 2.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 3.7 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 4.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 4.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 1.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 1.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 2.0 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 2.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 12.3 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.1 | 12.2 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.1 | 4.6 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 5.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 17.1 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.1 | 2.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 3.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.6 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 3.1 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 1.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 2.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 36.4 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 5.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 3.1 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 4.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.9 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 1.0 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.1 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 9.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.9 | 7.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.6 | 10.7 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.5 | 10.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.5 | 6.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.3 | 11.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 2.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.2 | 6.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.2 | 5.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.2 | 5.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.7 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 11.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.3 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 10.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 1.1 | 9.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 1.0 | 7.8 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.7 | 6.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.6 | 4.1 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.5 | 3.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.3 | 10.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 4.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 5.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.3 | 2.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 5.5 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.3 | 21.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.2 | 3.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.9 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 6.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 1.3 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 2.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 5.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.4 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |