Project
DANIO-CODE
Navigation
Downloads
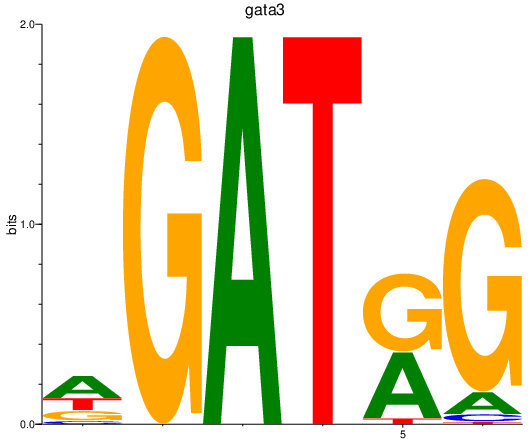
Results for gata3
Z-value: 1.62
Transcription factors associated with gata3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gata3
|
ENSDARG00000016526 | GATA binding protein 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gata3 | dr10_dc_chr4_-_25080470_25080483 | 0.80 | 2.2e-04 | Click! |
Activity profile of gata3 motif
Sorted Z-values of gata3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of gata3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_31490043 | 6.82 |
ENSDART00000076636
|
fzd2
|
frizzled class receptor 2 |
| chr12_-_5470187 | 6.37 |
ENSDART00000159784
|
abi3b
|
ABI family, member 3b |
| chr7_+_23778570 | 5.74 |
ENSDART00000121837
|
efs
|
embryonal Fyn-associated substrate |
| chr19_-_33123724 | 5.18 |
ENSDART00000004034
|
hpca
|
hippocalcin |
| chr11_-_471166 | 5.15 |
ENSDART00000154888
|
cnbpb
|
CCHC-type zinc finger, nucleic acid binding protein b |
| chr21_-_43083936 | 4.74 |
ENSDART00000040169
|
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr7_+_73548430 | 4.36 |
ENSDART00000092691
|
pea15
|
phosphoprotein enriched in astrocytes 15 |
| chr23_-_39956151 | 4.17 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr4_-_19895115 | 4.12 |
ENSDART00000105967
|
cacna2d1a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1a |
| chr2_+_51866323 | 3.86 |
ENSDART00000163644
|
zgc:165603
|
zgc:165603 |
| chr8_-_12365342 | 3.85 |
ENSDART00000113286
|
phf19
|
PHD finger protein 19 |
| chr7_-_24428781 | 3.83 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr16_-_29502741 | 3.71 |
ENSDART00000148405
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr14_-_31676235 | 3.69 |
ENSDART00000105761
|
zic3
|
zic family member 3 heterotaxy 1 (odd-paired homolog, Drosophila) |
| chr2_-_42643045 | 3.66 |
ENSDART00000139929
|
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr4_-_4923982 | 3.64 |
ENSDART00000103293
|
ndufa5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr8_+_8056616 | 3.47 |
|
|
|
| chr8_-_12365285 | 3.43 |
ENSDART00000142150
|
phf19
|
PHD finger protein 19 |
| chr7_-_69639922 | 3.43 |
|
|
|
| chr7_-_37350985 | 3.34 |
ENSDART00000173523
|
nkd1
|
naked cuticle homolog 1 (Drosophila) |
| chr4_-_19895001 | 3.30 |
ENSDART00000014440
|
cacna2d1a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1a |
| chr10_-_10059864 | 3.26 |
ENSDART00000132375
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr10_+_31701855 | 2.99 |
ENSDART00000115251
|
esama
|
endothelial cell adhesion molecule a |
| chr16_-_20506304 | 2.91 |
ENSDART00000134980
|
si:dkeyp-86h10.3
|
si:dkeyp-86h10.3 |
| chr19_+_12525407 | 2.83 |
ENSDART00000135706
|
ldlrad4a
|
low density lipoprotein receptor class A domain containing 4a |
| chr20_-_9474672 | 2.81 |
ENSDART00000152674
|
ENSDARG00000033201
|
ENSDARG00000033201 |
| chr6_+_27072030 | 2.74 |
ENSDART00000149429
|
boka
|
BCL2-related ovarian killer a |
| chr17_-_21260319 | 2.72 |
ENSDART00000166524
|
hspa12a
|
heat shock protein 12A |
| chr14_+_36156947 | 2.62 |
|
|
|
| chr17_-_48623315 | 2.60 |
ENSDART00000030934
|
kcnk5a
|
potassium channel, subfamily K, member 5a |
| chr5_-_65349550 | 2.57 |
ENSDART00000164228
|
nrarpb
|
notch-regulated ankyrin repeat protein b |
| chr25_+_17308788 | 2.44 |
ENSDART00000164349
|
e2f4
|
E2F transcription factor 4 |
| chr12_+_30538113 | 2.43 |
ENSDART00000179355
|
thbs2b
|
thrombospondin 2b |
| chr18_+_618393 | 2.40 |
ENSDART00000159464
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr9_+_52872108 | 2.37 |
ENSDART00000157453
ENSDART00000163684 |
nme8
|
NME/NM23 family member 8 |
| chr7_-_13638486 | 2.36 |
ENSDART00000169828
|
rlbp1a
|
retinaldehyde binding protein 1a |
| chr19_-_44223619 | 2.33 |
ENSDART00000160879
|
klhl43
|
kelch-like family member 43 |
| chr17_-_8016214 | 2.31 |
ENSDART00000148520
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr13_+_18414806 | 2.28 |
ENSDART00000110197
|
zgc:154058
|
zgc:154058 |
| chr2_-_42642885 | 2.28 |
ENSDART00000139929
|
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr14_+_6239755 | 2.24 |
ENSDART00000097214
|
adam19b
|
ADAM metallopeptidase domain 19b |
| chr12_+_5154886 | 2.22 |
ENSDART00000163897
|
lgi1b
|
leucine-rich, glioma inactivated 1b |
| chr14_+_45619218 | 2.18 |
ENSDART00000158723
|
sncb
|
synuclein, beta |
| chr12_+_36716775 | 2.18 |
ENSDART00000048927
|
cox10
|
COX10 heme A:farnesyltransferase cytochrome c oxidase assembly factor |
| chr18_-_44135616 | 2.16 |
ENSDART00000087339
|
cdon
|
cell adhesion associated, oncogene regulated |
| KN150698v1_+_1663 | 2.11 |
ENSDART00000163899
|
ENSDARG00000100052
|
ENSDARG00000100052 |
| chr16_+_38335706 | 2.09 |
|
|
|
| chr6_+_54134129 | 2.07 |
ENSDART00000156554
|
hmga1b
|
high mobility group AT-hook 1b |
| chr24_+_27469268 | 2.07 |
ENSDART00000105774
|
ek1
|
eph-like kinase 1 |
| chr2_+_48448974 | 2.04 |
ENSDART00000023040
|
hes6
|
hes family bHLH transcription factor 6 |
| chr2_+_35871754 | 2.01 |
ENSDART00000134918
|
dhx9
|
DEAH (Asp-Glu-Ala-His) box helicase 9 |
| chr19_+_3713027 | 1.99 |
ENSDART00000125673
|
nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr19_-_41482388 | 1.97 |
ENSDART00000111982
|
sgce
|
sarcoglycan, epsilon |
| chr18_+_6551983 | 1.93 |
ENSDART00000160382
ENSDART00000171495 ENSDART00000160228 |
fam168a
|
family with sequence similarity 168, member A |
| chr19_-_33123907 | 1.92 |
ENSDART00000004034
|
hpca
|
hippocalcin |
| chr18_-_46356213 | 1.87 |
ENSDART00000010813
|
foxa3
|
forkhead box A3 |
| chr20_-_4723724 | 1.86 |
ENSDART00000147071
|
ak7a
|
adenylate kinase 7a |
| chr11_+_34562255 | 1.85 |
ENSDART00000103157
|
slc38a3a
|
solute carrier family 38, member 3a |
| chr5_-_2014043 | 1.84 |
ENSDART00000064012
|
ca4a
|
carbonic anhydrase IV a |
| chr8_+_39573104 | 1.81 |
ENSDART00000158498
ENSDART00000041634 ENSDART00000169542 |
msi1
|
musashi RNA-binding protein 1 |
| chr15_+_14048904 | 1.80 |
ENSDART00000159438
|
zgc:162730
|
zgc:162730 |
| chr17_-_43725003 | 1.78 |
|
|
|
| chr21_-_1716495 | 1.75 |
ENSDART00000151049
|
onecut2
|
one cut homeobox 2 |
| chr20_+_47937533 | 1.72 |
ENSDART00000159983
|
CABZ01059119.1
|
ENSDARG00000099804 |
| chr8_+_8006488 | 1.72 |
|
|
|
| chr2_+_21342233 | 1.72 |
ENSDART00000062563
|
rreb1b
|
ras responsive element binding protein 1b |
| chr15_-_15421065 | 1.70 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr7_+_73466424 | 1.69 |
ENSDART00000123429
|
si:dkey-46i9.6
|
si:dkey-46i9.6 |
| chr10_+_19069375 | 1.69 |
ENSDART00000111952
|
igsf9a
|
immunoglobulin superfamily, member 9a |
| chr3_-_21111908 | 1.69 |
ENSDART00000007293
|
tcap
|
titin-cap (telethonin) |
| chr8_-_6782029 | 1.65 |
ENSDART00000135834
ENSDART00000172157 |
dock5
|
dedicator of cytokinesis 5 |
| chr20_+_18263793 | 1.65 |
ENSDART00000011287
|
aqp4
|
aquaporin 4 |
| chr3_+_18406137 | 1.64 |
ENSDART00000158791
|
cbx4
|
chromobox homolog 4 (Pc class homolog, Drosophila) |
| chr25_-_10992022 | 1.60 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr19_+_31474352 | 1.59 |
ENSDART00000145971
|
sostdc1b
|
sclerostin domain containing 1b |
| chr15_+_28753020 | 1.58 |
ENSDART00000155815
|
nova2
|
neuro-oncological ventral antigen 2 |
| chr17_+_32390603 | 1.57 |
ENSDART00000156051
|
dhx32b
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32b |
| chr9_+_48717490 | 1.56 |
ENSDART00000163353
|
lrp2a
|
low density lipoprotein receptor-related protein 2a |
| chr14_+_6834563 | 1.54 |
ENSDART00000170994
ENSDART00000129898 |
ctsf
|
cathepsin F |
| chr4_-_12719490 | 1.49 |
ENSDART00000035259
|
mgst1.1
|
microsomal glutathione S-transferase 1.1 |
| chr3_+_26013873 | 1.49 |
ENSDART00000043932
|
atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr20_-_40217185 | 1.45 |
|
|
|
| chr18_-_22764560 | 1.43 |
ENSDART00000009912
|
hsf4
|
heat shock transcription factor 4 |
| chr13_-_31413855 | 1.43 |
ENSDART00000005670
|
dhrs7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr7_+_73548348 | 1.43 |
ENSDART00000092691
|
pea15
|
phosphoprotein enriched in astrocytes 15 |
| chr15_+_40161311 | 1.41 |
ENSDART00000063783
|
itm2ca
|
integral membrane protein 2Ca |
| chr3_-_55956030 | 1.41 |
ENSDART00000158163
|
srl
|
sarcalumenin |
| chr17_-_23235070 | 1.41 |
|
|
|
| chr23_-_14317229 | 1.39 |
|
|
|
| chr5_+_71028018 | 1.37 |
ENSDART00000164893
ENSDART00000159658 ENSDART00000097164 ENSDART00000124939 ENSDART00000171230 |
LHX3
|
LIM homeobox 3 |
| chr20_-_35938683 | 1.35 |
ENSDART00000037855
|
tnfrsf21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr25_+_33527870 | 1.34 |
ENSDART00000011967
|
anxa2a
|
annexin A2a |
| chr15_+_41858271 | 1.33 |
ENSDART00000059508
|
pxylp1
|
2-phosphoxylose phosphatase 1 |
| chr11_+_1601946 | 1.33 |
ENSDART00000154967
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr4_+_11312432 | 1.33 |
ENSDART00000051792
|
sema3aa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Aa |
| chr5_+_51392394 | 1.31 |
ENSDART00000143276
|
setbp1
|
SET binding protein 1 |
| chr24_+_389982 | 1.31 |
ENSDART00000061973
|
tgfbr1b
|
transforming growth factor, beta receptor 1 b |
| chr6_+_27071986 | 1.26 |
ENSDART00000149429
|
boka
|
BCL2-related ovarian killer a |
| chr17_-_37266525 | 1.26 |
ENSDART00000128715
|
kif3cb
|
kinesin family member 3Cb |
| chr19_+_26838851 | 1.25 |
ENSDART00000136244
|
npr1a
|
natriuretic peptide receptor 1a |
| chr11_-_40255633 | 1.22 |
ENSDART00000172819
|
miip
|
migration and invasion inhibitory protein |
| chr9_-_30463616 | 1.21 |
ENSDART00000089526
|
otc
|
ornithine carbamoyltransferase |
| chr24_+_40974634 | 1.21 |
|
|
|
| chr4_-_25869361 | 1.20 |
ENSDART00000128968
|
nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr20_-_51746555 | 1.20 |
ENSDART00000065231
|
disp1
|
dispatched homolog 1 (Drosophila) |
| chr17_+_45834257 | 1.19 |
ENSDART00000035152
|
kif26ab
|
kinesin family member 26Ab |
| chr10_+_42761097 | 1.19 |
ENSDART00000075259
|
fgfr1b
|
fibroblast growth factor receptor 1b |
| chr1_-_5527494 | 1.18 |
ENSDART00000131254
|
mdh1b
|
malate dehydrogenase 1B, NAD (soluble) |
| chr13_+_1314139 | 1.18 |
|
|
|
| chr25_+_31547276 | 1.17 |
ENSDART00000090727
|
duox
|
dual oxidase |
| chr16_-_1479139 | 1.14 |
ENSDART00000036348
|
sim1a
|
single-minded family bHLH transcription factor 1a |
| chr7_+_58718614 | 1.13 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr15_-_25592228 | 1.13 |
ENSDART00000157498
|
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr23_-_43595189 | 1.13 |
|
|
|
| chr8_+_2517472 | 1.12 |
ENSDART00000112914
|
si:ch211-51h9.7
|
si:ch211-51h9.7 |
| chr8_-_14142049 | 1.12 |
ENSDART00000126432
|
rhoaa
|
ras homolog gene family, member Aa |
| chr6_-_39767452 | 1.11 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr1_-_22867130 | 1.09 |
ENSDART00000145942
ENSDART00000077394 |
fam184b
|
family with sequence similarity 184, member B |
| chr6_+_29800606 | 1.09 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr4_-_22509152 | 1.06 |
ENSDART00000176950
|
ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr17_+_15025997 | 1.05 |
ENSDART00000154987
|
samd4a
|
sterile alpha motif domain containing 4A |
| chr10_-_10372266 | 1.01 |
|
|
|
| chr21_+_8106096 | 1.01 |
ENSDART00000011096
|
nr6a1b
|
nuclear receptor subfamily 6, group A, member 1b |
| chr13_+_35619748 | 1.01 |
ENSDART00000159690
|
gpr75
|
G protein-coupled receptor 75 |
| chr21_+_10983499 | 0.99 |
ENSDART00000039549
|
prlra
|
prolactin receptor a |
| chr9_-_7695437 | 0.98 |
ENSDART00000102715
|
tuba8l3
|
tubulin, alpha 8 like 3 |
| chr11_+_40945681 | 0.97 |
|
|
|
| chr8_-_8407754 | 0.97 |
ENSDART00000137382
|
cdk16
|
cyclin-dependent kinase 16 |
| chr10_+_33935945 | 0.97 |
|
|
|
| chr5_+_31606213 | 0.97 |
ENSDART00000126873
|
myhz2
|
myosin, heavy polypeptide 2, fast muscle specific |
| chr22_-_23241074 | 0.96 |
ENSDART00000175556
|
lhx9
|
LIM homeobox 9 |
| chr18_-_21058499 | 0.96 |
ENSDART00000010189
|
igf1ra
|
insulin-like growth factor 1a receptor |
| chr4_-_2497857 | 0.96 |
ENSDART00000058059
|
OSBPL8
|
oxysterol binding protein like 8 |
| chr18_-_48750637 | 0.95 |
ENSDART00000097259
|
st3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr3_+_16115708 | 0.95 |
ENSDART00000122519
|
st8sia6
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
| chr10_+_13042496 | 0.94 |
ENSDART00000158919
|
lpar1
|
lysophosphatidic acid receptor 1 |
| chr6_-_43094573 | 0.94 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr15_+_28370102 | 0.93 |
ENSDART00000175860
|
myo1cb
|
myosin Ic, paralog b |
| chr16_-_29502678 | 0.93 |
ENSDART00000148405
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr9_+_30483319 | 0.92 |
ENSDART00000148148
|
srpx
|
sushi-repeat containing protein, X-linked |
| chr1_-_51494196 | 0.90 |
ENSDART00000149939
|
rad23aa
|
RAD23 homolog A, nucleotide excision repair protein a |
| chr4_+_6634849 | 0.90 |
ENSDART00000099462
|
gpr85
|
G protein-coupled receptor 85 |
| chr3_+_23090404 | 0.90 |
ENSDART00000009393
|
col1a1a
|
collagen, type I, alpha 1a |
| chr8_+_41639900 | 0.89 |
ENSDART00000136492
|
si:ch211-158d24.4
|
si:ch211-158d24.4 |
| chr1_+_25717717 | 0.87 |
ENSDART00000112263
|
arhgef38
|
Rho guanine nucleotide exchange factor (GEF) 38 |
| chr5_-_11443522 | 0.87 |
ENSDART00000074979
|
rnft2
|
ring finger protein, transmembrane 2 |
| chr5_+_16669696 | 0.87 |
|
|
|
| chr4_-_4590314 | 0.87 |
ENSDART00000082070
|
rassf3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr23_+_36010592 | 0.86 |
ENSDART00000137507
|
hoxc3a
|
homeo box C3a |
| chr12_-_25973094 | 0.86 |
ENSDART00000171206
ENSDART00000171212 ENSDART00000170265 |
ldb3b
|
LIM domain binding 3b |
| chr10_-_20489955 | 0.86 |
ENSDART00000159672
|
loxl2a
|
lysyl oxidase-like 2a |
| chr23_-_33018067 | 0.86 |
|
|
|
| chr15_+_23849554 | 0.85 |
ENSDART00000138375
|
ift20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr19_-_43037791 | 0.85 |
ENSDART00000004392
|
fkbp9
|
FK506 binding protein 9 |
| chr12_+_22701981 | 0.85 |
ENSDART00000153199
|
afap1
|
actin filament associated protein 1 |
| chr25_+_31746643 | 0.85 |
ENSDART00000162636
|
tjp1b
|
tight junction protein 1b |
| chr16_+_23172295 | 0.84 |
ENSDART00000167518
ENSDART00000161087 |
efna3b
|
ephrin-A3b |
| chr21_-_11539798 | 0.84 |
ENSDART00000144770
|
cast
|
calpastatin |
| chr20_+_33391554 | 0.84 |
ENSDART00000024104
|
mycn
|
MYCN proto-oncogene, bHLH transcription factor |
| chr20_+_22780756 | 0.83 |
|
|
|
| chr10_+_25304728 | 0.83 |
ENSDART00000100438
|
rab38b
|
RAB38b, member of RAS oncogene family |
| chr2_-_52202004 | 0.83 |
ENSDART00000165350
|
BX908782.1
|
ENSDARG00000098957 |
| chr15_+_7179382 | 0.83 |
ENSDART00000101578
|
her8.2
|
hairy-related 8.2 |
| chr18_-_34166500 | 0.82 |
ENSDART00000079341
|
plch1
|
phospholipase C, eta 1 |
| chr17_+_15026344 | 0.82 |
ENSDART00000154987
|
samd4a
|
sterile alpha motif domain containing 4A |
| chr4_-_25496215 | 0.82 |
ENSDART00000041402
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr6_+_7936632 | 0.81 |
ENSDART00000146106
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr5_+_15167637 | 0.81 |
ENSDART00000127015
|
srrm4
|
serine/arginine repetitive matrix 4 |
| chr18_+_24932972 | 0.81 |
ENSDART00000008638
|
rgma
|
repulsive guidance molecule family member a |
| chr1_-_50147413 | 0.81 |
ENSDART00000080389
|
fam13a
|
family with sequence similarity 13, member A |
| chr7_-_26767182 | 0.81 |
ENSDART00000052731
|
nucb2a
|
nucleobindin 2a |
| chr5_-_35729429 | 0.80 |
|
|
|
| chr13_-_28543929 | 0.80 |
ENSDART00000122754
ENSDART00000057574 |
nt5c2a
|
5'-nucleotidase, cytosolic IIa |
| chr12_-_33792377 | 0.79 |
ENSDART00000153280
|
sema4gb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Gb |
| chr25_+_24193604 | 0.79 |
ENSDART00000083407
|
b4galnt4a
|
beta-1,4-N-acetyl-galactosaminyl transferase 4a |
| chr7_-_43501747 | 0.79 |
ENSDART00000002279
|
cdh11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr13_+_11913290 | 0.79 |
ENSDART00000079398
|
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr6_-_49160398 | 0.79 |
ENSDART00000143241
|
tspan2a
|
tetraspanin 2a |
| chr8_+_2380040 | 0.79 |
ENSDART00000141263
|
ENKD1
|
enkurin domain containing 1 |
| chr2_-_31037043 | 0.78 |
ENSDART00000087026
|
emilin2a
|
elastin microfibril interfacer 2a |
| chr8_+_23071884 | 0.78 |
ENSDART00000063075
|
zgc:100920
|
zgc:100920 |
| chr18_+_49230636 | 0.77 |
ENSDART00000167609
ENSDART00000135026 ENSDART00000171618 |
si:ch211-136a13.1
|
si:ch211-136a13.1 |
| chr23_-_35595271 | 0.77 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr10_-_13814731 | 0.77 |
ENSDART00000145103
|
cntfr
|
ciliary neurotrophic factor receptor |
| chr5_+_8460247 | 0.77 |
ENSDART00000091397
|
tnksb
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase b |
| chr15_+_6070537 | 0.75 |
ENSDART00000123797
|
pcp4b
|
Purkinje cell protein 4b |
| chr23_+_20183765 | 0.75 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| KN150256v1_-_9467 | 0.75 |
|
|
|
| chr17_+_31722419 | 0.74 |
ENSDART00000155073
|
arhgap5
|
Rho GTPase activating protein 5 |
| chr1_+_54231447 | 0.74 |
ENSDART00000145652
|
golga7ba
|
golgin A7 family, member Ba |
| chr1_-_20131454 | 0.74 |
|
|
|
| chr1_+_35462641 | 0.74 |
ENSDART00000053773
ENSDART00000147458 |
lsm6
|
LSM6 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr10_+_1812271 | 0.74 |
|
|
|
| chr23_-_38228305 | 0.74 |
ENSDART00000131791
|
pfdn4
|
prefoldin subunit 4 |
| chr14_+_20809272 | 0.74 |
ENSDART00000139865
|
aldob
|
aldolase b, fructose-bisphosphate |
| chr22_+_15412270 | 0.73 |
ENSDART00000010846
|
gpc5b
|
glypican 5b |
| chr15_-_31687 | 0.73 |
|
|
|
| chr4_-_18466436 | 0.73 |
ENSDART00000150221
|
sco2
|
SCO2 cytochrome c oxidase assembly protein |
| chr11_+_34788205 | 0.73 |
ENSDART00000124800
|
fam212aa
|
family with sequence similarity 212, member Aa |
| chr18_+_30391910 | 0.73 |
ENSDART00000158871
|
gse1
|
Gse1 coiled-coil protein |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.7 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 1.3 | 3.8 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.9 | 5.2 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.8 | 3.3 | GO:1903533 | regulation of protein targeting(GO:1903533) |
| 0.7 | 3.5 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.6 | 7.5 | GO:0003306 | Wnt signaling pathway involved in heart development(GO:0003306) |
| 0.5 | 2.1 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.5 | 1.5 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) positive regulation of muscle contraction(GO:0045933) |
| 0.5 | 1.9 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.4 | 1.3 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.4 | 1.3 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.4 | 1.3 | GO:0097377 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.4 | 1.7 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.4 | 1.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.4 | 1.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.4 | 1.2 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.4 | 1.8 | GO:0015817 | histidine transport(GO:0015817) |
| 0.3 | 2.4 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.3 | 1.6 | GO:0006833 | water transport(GO:0006833) |
| 0.3 | 1.2 | GO:0000050 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.3 | 0.9 | GO:0070977 | ossification involved in bone maturation(GO:0043931) bone maturation(GO:0070977) |
| 0.3 | 3.0 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.3 | 2.9 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.3 | 1.1 | GO:0035776 | pronephric proximal tubule development(GO:0035776) |
| 0.3 | 0.8 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.3 | 1.4 | GO:0010561 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.3 | 0.3 | GO:0014829 | vascular smooth muscle contraction(GO:0014829) |
| 0.3 | 0.8 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.3 | 2.4 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.3 | 2.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.3 | 1.6 | GO:0070293 | renal absorption(GO:0070293) |
| 0.3 | 0.8 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.2 | 1.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.2 | 0.9 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.2 | 0.9 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.2 | 2.6 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.2 | 0.6 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.2 | 1.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.2 | 0.2 | GO:0010595 | positive regulation of endothelial cell migration(GO:0010595) |
| 0.2 | 0.9 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.2 | 0.5 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.2 | 0.5 | GO:1902871 | negative regulation of photoreceptor cell differentiation(GO:0046533) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.2 | 0.9 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 1.2 | GO:0006735 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 1.3 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.2 | 0.8 | GO:0090104 | pancreatic epsilon cell differentiation(GO:0090104) |
| 0.2 | 0.3 | GO:0050886 | endocrine process(GO:0050886) |
| 0.2 | 1.0 | GO:0032096 | regulation of response to food(GO:0032095) negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.2 | 2.1 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 1.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.9 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.1 | 3.4 | GO:0038034 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.1 | 0.6 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.1 | 0.4 | GO:0042671 | blood vessel maturation(GO:0001955) retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 0.5 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.4 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.1 | 0.9 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.1 | 2.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 6.8 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 1.2 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.1 | 0.5 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 0.4 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 0.6 | GO:1901739 | regulation of myoblast fusion(GO:1901739) |
| 0.1 | 2.3 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 1.0 | GO:0014831 | gastro-intestinal system smooth muscle contraction(GO:0014831) |
| 0.1 | 0.4 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.1 | 2.1 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.1 | 0.2 | GO:0030852 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.1 | 0.9 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.1 | 0.8 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 0.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.7 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 2.4 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 0.2 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.1 | 2.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 2.1 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.1 | 0.4 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.1 | 0.2 | GO:0060402 | cytosolic calcium ion transport(GO:0060401) calcium ion transport into cytosol(GO:0060402) |
| 0.1 | 0.8 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 9.6 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.1 | 1.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.8 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 0.6 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.1 | 0.5 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.4 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.1 | 0.2 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 0.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 1.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.3 | GO:0003379 | establishment of cell polarity involved in gastrulation cell migration(GO:0003379) |
| 0.1 | 0.3 | GO:1904969 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.1 | 0.2 | GO:0044036 | cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.1 | 0.5 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 1.0 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.1 | 2.9 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.1 | 3.5 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.1 | 0.4 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.1 | 0.2 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 1.0 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 | 0.3 | GO:0048790 | neuromuscular synaptic transmission(GO:0007274) maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.1 | 1.0 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 6.0 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.1 | 2.2 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.1 | 1.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.5 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.1 | 0.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.4 | GO:0003313 | heart rudiment development(GO:0003313) |
| 0.1 | 0.5 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.6 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 0.6 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 1.4 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.1 | 0.3 | GO:0019878 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.1 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.8 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 0.4 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.1 | 0.4 | GO:0021576 | hindbrain formation(GO:0021576) |
| 0.1 | 0.3 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.1 | 0.2 | GO:0015867 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 0.6 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.1 | 0.4 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.3 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.5 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.1 | 0.5 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 0.5 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 0.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.2 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) |
| 0.0 | 0.1 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.3 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 1.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.5 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.2 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.4 | GO:0043011 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) dendritic cell differentiation(GO:0097028) |
| 0.0 | 2.4 | GO:0006165 | nucleoside diphosphate phosphorylation(GO:0006165) |
| 0.0 | 0.2 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:0002761 | regulation of myeloid leukocyte differentiation(GO:0002761) |
| 0.0 | 0.4 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.1 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.4 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.6 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.3 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.4 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.3 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.2 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.0 | 0.8 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.3 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 1.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.4 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.3 | GO:0098926 | postsynaptic signal transduction(GO:0098926) |
| 0.0 | 0.5 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) |
| 0.0 | 0.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.2 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.0 | 0.4 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.5 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.5 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.7 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.6 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 3.5 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.4 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 1.6 | GO:1901342 | regulation of vasculature development(GO:1901342) |
| 0.0 | 0.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.4 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 1.9 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0042537 | benzene-containing compound metabolic process(GO:0042537) |
| 0.0 | 0.1 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.6 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.2 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 0.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.4 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 1.3 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 4.7 | GO:0023014 | MAPK cascade(GO:0000165) signal transduction by protein phosphorylation(GO:0023014) |
| 0.0 | 0.5 | GO:0021986 | habenula development(GO:0021986) |
| 0.0 | 0.2 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 1.3 | GO:0032835 | glomerulus development(GO:0032835) |
| 0.0 | 0.3 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.1 | GO:0055004 | atrial cardiac myofibril assembly(GO:0055004) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.3 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 1.4 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.1 | GO:1901909 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.3 | GO:2000369 | positive regulation of Notch signaling pathway(GO:0045747) regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.7 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.2 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) |
| 0.0 | 0.3 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 1.2 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.6 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.2 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.8 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.2 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.4 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 1.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.9 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.1 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) negative regulation of histone ubiquitination(GO:0033183) histone H2A K63-linked ubiquitination(GO:0070535) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.2 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.5 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.0 | 0.1 | GO:0032108 | negative regulation of response to extracellular stimulus(GO:0032105) negative regulation of response to nutrient levels(GO:0032108) |
| 0.0 | 0.8 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.0 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.4 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.4 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.0 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.5 | 7.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.4 | 1.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.3 | 2.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 6.0 | GO:0043679 | axon terminus(GO:0043679) |
| 0.2 | 2.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.2 | 6.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 1.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.7 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 1.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 2.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.9 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 0.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.5 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.5 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.1 | 3.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 6.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 0.7 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.8 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.7 | GO:0044447 | axoneme part(GO:0044447) |
| 0.1 | 1.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.7 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 1.0 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.3 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 4.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 0.8 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 2.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 1.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 2.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 2.3 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 2.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.2 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.6 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 3.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 2.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 1.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.4 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 2.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.8 | GO:0060293 | germ plasm(GO:0060293) |
| 0.0 | 6.8 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 1.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.2 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 26.3 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.2 | GO:0071256 | translocon complex(GO:0071256) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.7 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0098594 | mucin granule(GO:0098594) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.6 | GO:0031968 | outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.0 | 0.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.5 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 1.7 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.7 | 2.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.6 | 2.9 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.5 | 1.9 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.5 | 1.8 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) L-asparagine transmembrane transporter activity(GO:0015182) |
| 0.4 | 1.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.4 | 1.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.4 | 3.7 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.4 | 1.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.4 | 1.2 | GO:0016175 | NAD(P)H oxidase activity(GO:0016174) superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.4 | 1.1 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.4 | 6.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 4.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.3 | 1.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.3 | 0.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.3 | 8.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.3 | 1.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 1.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.3 | 1.6 | GO:0036122 | BMP binding(GO:0036122) |
| 0.3 | 0.8 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.2 | 0.7 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.2 | 7.1 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.2 | 3.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 0.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 2.3 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.2 | 2.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 0.9 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.2 | 1.2 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 1.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.2 | 0.7 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 8.4 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 2.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 7.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.0 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.9 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 5.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.5 | GO:0019809 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.1 | 0.5 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 1.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.5 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 1.0 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.1 | 1.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 0.3 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.1 | 1.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.3 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 1.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.8 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 1.6 | GO:0042562 | hormone binding(GO:0042562) |
| 0.1 | 2.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 0.4 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.1 | 0.3 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.5 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 0.3 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.1 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 1.5 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 0.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.2 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.1 | 1.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 3.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 3.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 1.4 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.1 | 3.5 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.1 | 0.8 | GO:0008443 | phosphofructokinase activity(GO:0008443) |
| 0.1 | 0.4 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.2 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.1 | 0.4 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.2 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 0.3 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.1 | 0.6 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.1 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 1.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 1.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.9 | GO:0042805 | actinin binding(GO:0042805) muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 1.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 0.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.4 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 24.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 1.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.0 | 0.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.4 | GO:0097200 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.6 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) neurotrophin binding(GO:0043121) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.3 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 6.2 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 1.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 1.6 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 2.3 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.1 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.3 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.5 | GO:0036442 | hydrogen-exporting ATPase activity(GO:0036442) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 2.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.9 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.1 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.2 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.4 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 1.8 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.4 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 21.3 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 2.0 | GO:0019900 | kinase binding(GO:0019900) |
| 0.0 | 0.7 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 2.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.6 | GO:0003779 | actin binding(GO:0003779) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.0 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 0.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 2.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 3.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 6.5 | PID P73PATHWAY | p73 transcription factor network |
| 0.1 | 0.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 2.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 3.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 2.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 1.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 0.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 2.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.9 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.7 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.3 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.1 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.1 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 8.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.2 | 1.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.2 | 0.5 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 1.6 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.1 | 1.9 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 0.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 1.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 0.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 1.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 4.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 0.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.2 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 0.5 | REACTOME METABOLISM OF RNA | Genes involved in Metabolism of RNA |
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 2.0 | REACTOME PROTEIN FOLDING | Genes involved in Protein folding |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME GPCR LIGAND BINDING | Genes involved in GPCR ligand binding |
| 0.0 | 1.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |