Project
DANIO-CODE
Navigation
Downloads
Results for gata4
Z-value: 0.45
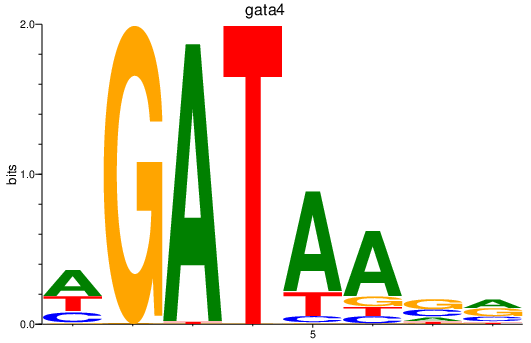
Transcription factors associated with gata4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gata4
|
ENSDARG00000098952 | GATA binding protein 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gata4 | dr10_dc_chr20_-_53162473_53162571 | 0.33 | 2.1e-01 | Click! |
Activity profile of gata4 motif
Sorted Z-values of gata4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of gata4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_25157675 | 0.79 |
ENSDART00000136984
|
fabp2
|
fatty acid binding protein 2, intestinal |
| chr10_-_32614936 | 0.70 |
ENSDART00000143301
|
mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr15_-_18425091 | 0.65 |
ENSDART00000155866
|
zbtb16b
|
zinc finger and BTB domain containing 16b |
| chr3_+_32360862 | 0.55 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr8_-_7049946 | 0.46 |
ENSDART00000045669
ENSDART00000162950 |
KRT73
|
keratin 73 |
| chr25_+_21732255 | 0.41 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr2_-_47578695 | 0.39 |
ENSDART00000014350
ENSDART00000038828 |
pax3a
|
paired box 3a |
| chr12_+_6007990 | 0.37 |
ENSDART00000091868
|
g6pca.2
|
glucose-6-phosphatase a, catalytic subunit, tandem duplicate 2 |
| chr10_+_10843384 | 0.36 |
ENSDART00000130283
|
ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr3_+_12613413 | 0.35 |
ENSDART00000166071
|
cyp2k16
|
cytochrome P450, family 2, subfamily K, polypeptide16 |
| chr21_-_40811269 | 0.35 |
ENSDART00000148513
|
limk1b
|
LIM domain kinase 1b |
| chr11_+_21749658 | 0.34 |
ENSDART00000161485
|
foxp4
|
forkhead box P4 |
| chr8_-_41194452 | 0.33 |
ENSDART00000165949
|
fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr9_+_17298410 | 0.33 |
ENSDART00000048548
|
scel
|
sciellin |
| chr13_-_30515486 | 0.31 |
ENSDART00000139607
|
zcchc24
|
zinc finger, CCHC domain containing 24 |
| chr8_-_50299273 | 0.31 |
ENSDART00000023639
|
nkx2.7
|
NK2 transcription factor related 7 |
| chr7_+_34018862 | 0.29 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr20_-_25063937 | 0.29 |
ENSDART00000159122
|
epha7
|
eph receptor A7 |
| chr16_+_53238110 | 0.29 |
ENSDART00000102170
|
CABZ01053976.1
|
ENSDARG00000069929 |
| chr25_+_18467217 | 0.26 |
ENSDART00000170841
|
cav1
|
caveolin 1 |
| chr5_+_37996437 | 0.26 |
ENSDART00000015136
|
mogat3b
|
monoacylglycerol O-acyltransferase 3b |
| chr16_+_17061069 | 0.25 |
ENSDART00000111074
|
si:ch211-120k19.1
|
si:ch211-120k19.1 |
| chr11_+_31022048 | 0.25 |
ENSDART00000160154
|
si:dkey-238i5.2
|
si:dkey-238i5.2 |
| chr13_+_51154848 | 0.24 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr13_+_35213326 | 0.24 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr8_-_17580655 | 0.23 |
|
|
|
| chr8_+_23464087 | 0.23 |
ENSDART00000026316
|
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr23_-_20402258 | 0.23 |
ENSDART00000136204
|
CR749762.2
|
ENSDARG00000093223 |
| chr17_+_20549925 | 0.23 |
ENSDART00000113936
|
ENSDARG00000074663
|
ENSDARG00000074663 |
| chr4_-_21931540 | 0.23 |
ENSDART00000174400
|
rps16
|
ribosomal protein S16 |
| KN149726v1_+_1094 | 0.22 |
|
|
|
| chr3_+_28371725 | 0.22 |
ENSDART00000151081
|
sept12
|
septin 12 |
| chr6_+_2849460 | 0.20 |
|
|
|
| chr5_+_37996820 | 0.20 |
ENSDART00000015136
|
mogat3b
|
monoacylglycerol O-acyltransferase 3b |
| chr20_-_22478716 | 0.19 |
ENSDART00000110967
ENSDART00000011135 |
kita
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog a |
| chr21_-_9313788 | 0.19 |
|
|
|
| chr25_-_3730472 | 0.19 |
|
|
|
| chr18_+_18011562 | 0.19 |
ENSDART00000005027
|
ENSDARG00000011498
|
ENSDARG00000011498 |
| chr6_+_43236843 | 0.19 |
ENSDART00000112474
|
arl6ip5a
|
ADP-ribosylation factor-like 6 interacting protein 5a |
| chr16_+_23172295 | 0.18 |
ENSDART00000167518
ENSDART00000161087 |
efna3b
|
ephrin-A3b |
| chr1_-_11707288 | 0.18 |
ENSDART00000134708
|
sclt1
|
sodium channel and clathrin linker 1 |
| chr24_-_26328750 | 0.18 |
|
|
|
| chr12_-_36138709 | 0.18 |
ENSDART00000130985
|
st6galnac
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase |
| chr2_+_42286350 | 0.17 |
ENSDART00000008268
|
cyp7b1
|
cytochrome P450, family 7, subfamily B, polypeptide 1 |
| chr14_+_23673541 | 0.17 |
ENSDART00000124944
|
kif3a
|
kinesin family member 3A |
| chr6_-_43892838 | 0.17 |
ENSDART00000148646
ENSDART00000148909 |
foxp1b
|
forkhead box P1b |
| chr12_-_44825493 | 0.17 |
ENSDART00000016635
|
bccip
|
BRCA2 and CDKN1A interacting protein |
| chr16_+_24069711 | 0.16 |
ENSDART00000142869
|
apoc2
|
apolipoprotein C-II |
| chr20_-_21829447 | 0.16 |
|
|
|
| chr11_+_27970922 | 0.16 |
ENSDART00000169360
ENSDART00000043756 |
ephb2b
|
eph receptor B2b |
| chr23_-_30114528 | 0.15 |
ENSDART00000131209
|
ccdc187
|
coiled-coil domain containing 187 |
| chr16_+_21090083 | 0.15 |
ENSDART00000052662
|
hoxa13b
|
homeobox A13b |
| chr18_+_22228682 | 0.15 |
ENSDART00000165464
|
fam65a
|
family with sequence similarity 65, member A |
| chr6_-_16267366 | 0.15 |
ENSDART00000089445
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr12_+_15624640 | 0.15 |
ENSDART00000079803
|
nmt1b
|
N-myristoyltransferase 1b |
| chr16_+_24818799 | 0.14 |
ENSDART00000155217
|
si:dkey-79d12.4
|
si:dkey-79d12.4 |
| chr20_+_25567046 | 0.14 |
ENSDART00000153071
|
pfas
|
phosphoribosylformylglycinamidine synthase |
| chr22_-_20101177 | 0.14 |
ENSDART00000138688
|
creb3l3a
|
cAMP responsive element binding protein 3-like 3a |
| chr6_-_54103765 | 0.14 |
ENSDART00000083880
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr20_+_19093073 | 0.14 |
ENSDART00000147105
|
tdh
|
L-threonine dehydrogenase |
| chr2_-_6545431 | 0.13 |
ENSDART00000161934
|
si:dkey-119f1.1
|
si:dkey-119f1.1 |
| chr17_-_14663139 | 0.13 |
ENSDART00000037371
|
ppp1r13ba
|
protein phosphatase 1, regulatory subunit 13Ba |
| chr1_+_11485480 | 0.13 |
ENSDART00000132560
|
stra6l
|
STRA6-like |
| chr7_-_15582208 | 0.13 |
ENSDART00000102363
|
rcn1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr2_+_42286232 | 0.13 |
ENSDART00000008268
|
cyp7b1
|
cytochrome P450, family 7, subfamily B, polypeptide 1 |
| chr16_-_13652300 | 0.13 |
ENSDART00000144140
|
BX323458.2
|
ENSDARG00000094614 |
| chr8_+_8934762 | 0.13 |
ENSDART00000133137
|
bcap31
|
B-cell receptor-associated protein 31 |
| chr8_+_7739964 | 0.13 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr10_+_4114961 | 0.13 |
|
|
|
| chr8_-_31597252 | 0.12 |
ENSDART00000018886
|
ghra
|
growth hormone receptor a |
| chr9_+_33167554 | 0.12 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr25_-_13631707 | 0.12 |
ENSDART00000169865
|
lcat
|
lecithin-cholesterol acyltransferase |
| chr23_+_42370612 | 0.12 |
ENSDART00000161812
|
cyp2aa9
|
cytochrome P450, family 2, subfamily AA, polypeptide 9 |
| chr8_-_214434 | 0.12 |
ENSDART00000168412
|
hsd17b4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr8_-_24194821 | 0.12 |
ENSDART00000177464
|
CT025875.1
|
ENSDARG00000106046 |
| chr18_-_12302111 | 0.12 |
ENSDART00000062216
|
nup205
|
nucleoporin 205 |
| chr15_+_14925920 | 0.12 |
|
|
|
| chr6_+_58921655 | 0.11 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr19_-_46058123 | 0.11 |
|
|
|
| chr20_-_10500595 | 0.11 |
ENSDART00000064112
|
glrx5
|
glutaredoxin 5 homolog (S. cerevisiae) |
| chr23_-_33783396 | 0.11 |
ENSDART00000130338
|
pou6f1
|
POU class 6 homeobox 1 |
| chr6_+_12999420 | 0.11 |
ENSDART00000089725
|
ino80db
|
INO80 complex subunit Db |
| chr8_+_2260519 | 0.11 |
ENSDART00000136743
|
ikbkb
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
| chr2_-_6127811 | 0.11 |
ENSDART00000013079
|
scp2a
|
sterol carrier protein 2a |
| chr15_-_5827067 | 0.10 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr10_+_25234636 | 0.10 |
ENSDART00000024514
|
chordc1b
|
cysteine and histidine-rich domain (CHORD) containing 1b |
| chr3_+_1338347 | 0.10 |
ENSDART00000092690
ENSDART00000165395 |
srebf2
|
sterol regulatory element binding transcription factor 2 |
| chr9_-_27581087 | 0.10 |
ENSDART00000135221
|
nepro
|
nucleolus and neural progenitor protein |
| chr7_+_39409060 | 0.10 |
ENSDART00000133420
|
tbc1d14
|
TBC1 domain family, member 14 |
| chr7_-_51186389 | 0.09 |
ENSDART00000174328
|
arhgap6
|
Rho GTPase activating protein 6 |
| chr3_+_36830369 | 0.09 |
ENSDART00000150917
|
ENSDARG00000070174
|
ENSDARG00000070174 |
| chr15_-_1858350 | 0.09 |
ENSDART00000082026
|
mmp28
|
matrix metallopeptidase 28 |
| chr17_+_19479310 | 0.09 |
ENSDART00000077804
|
slc22a15
|
solute carrier family 22, member 15 |
| chr22_+_17484410 | 0.09 |
ENSDART00000088419
|
march2
|
membrane-associated ring finger (C3HC4) 2 |
| chr18_-_11439124 | 0.09 |
|
|
|
| chr24_+_37937721 | 0.09 |
ENSDART00000129889
|
ift140
|
intraflagellar transport 140 homolog (Chlamydomonas) |
| chr15_-_21901138 | 0.09 |
ENSDART00000089953
|
sik2b
|
salt-inducible kinase 2b |
| chr6_-_37765778 | 0.09 |
ENSDART00000149068
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr16_-_5936637 | 0.08 |
ENSDART00000085678
|
trak1
|
trafficking protein, kinesin binding 1 |
| chr18_+_24932972 | 0.08 |
ENSDART00000008638
|
rgma
|
repulsive guidance molecule family member a |
| chr19_+_4996328 | 0.08 |
ENSDART00000165082
|
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr11_-_27253835 | 0.08 |
ENSDART00000065889
|
wnt7aa
|
wingless-type MMTV integration site family, member 7Aa |
| chr17_-_13072982 | 0.08 |
ENSDART00000178859
|
CABZ01047998.1
|
ENSDARG00000105945 |
| chr20_+_6600246 | 0.08 |
ENSDART00000136567
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr16_-_13652342 | 0.08 |
ENSDART00000144140
|
BX323458.2
|
ENSDARG00000094614 |
| chr3_-_7220340 | 0.08 |
|
|
|
| chr18_+_14665460 | 0.08 |
ENSDART00000133834
|
vps9d1
|
VPS9 domain containing 1 |
| chr15_-_23587330 | 0.08 |
ENSDART00000167246
|
hmbsb
|
hydroxymethylbilane synthase, b |
| chr8_+_23334921 | 0.08 |
ENSDART00000085361
ENSDART00000046460 ENSDART00000145062 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr2_+_38279454 | 0.08 |
ENSDART00000076478
|
dhrs1
|
dehydrogenase/reductase (SDR family) member 1 |
| chr5_+_8741824 | 0.08 |
ENSDART00000081772
|
susd1
|
sushi domain containing 1 |
| chr8_-_41194530 | 0.08 |
ENSDART00000173055
|
fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr13_-_30515304 | 0.08 |
ENSDART00000139607
|
zcchc24
|
zinc finger, CCHC domain containing 24 |
| chr18_+_13280623 | 0.08 |
ENSDART00000080709
|
plcg2
|
phospholipase C, gamma 2 |
| chr24_-_13204960 | 0.08 |
ENSDART00000134482
|
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr9_+_17964241 | 0.07 |
ENSDART00000149736
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr13_+_24132236 | 0.07 |
ENSDART00000088005
|
abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr3_-_18261177 | 0.07 |
ENSDART00000041842
|
ndufb10
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 |
| chr20_+_23643050 | 0.07 |
ENSDART00000168690
|
palld
|
palladin, cytoskeletal associated protein |
| chr8_-_18547147 | 0.07 |
ENSDART00000123917
ENSDART00000063518 |
tmem47
|
transmembrane protein 47 |
| chr15_+_19902697 | 0.07 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr16_+_55121164 | 0.07 |
ENSDART00000149264
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr3_+_25888520 | 0.07 |
ENSDART00000135389
|
foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr8_+_7974156 | 0.07 |
|
|
|
| chr2_-_42011586 | 0.07 |
ENSDART00000045763
|
keap1a
|
kelch-like ECH-associated protein 1a |
| chr19_-_27730419 | 0.06 |
ENSDART00000151047
|
sh3bp5lb
|
SH3-binding domain protein 5-like, b |
| chr14_+_21388983 | 0.06 |
ENSDART00000137589
|
ran
|
RAN, member RAS oncogene family |
| chr2_+_56946960 | 0.06 |
|
|
|
| chr21_+_10773736 | 0.06 |
ENSDART00000137858
|
alpk2
|
alpha-kinase 2 |
| chr4_+_16721264 | 0.06 |
ENSDART00000035899
|
pkp2
|
plakophilin 2 |
| chr8_+_37667135 | 0.06 |
|
|
|
| chr11_+_21749918 | 0.06 |
ENSDART00000161485
|
foxp4
|
forkhead box P4 |
| chr17_+_21867968 | 0.06 |
ENSDART00000131929
|
plekha1a
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1a |
| chr13_+_33173742 | 0.06 |
ENSDART00000145295
|
dcdc2b
|
doublecortin domain containing 2B |
| chr13_+_18235298 | 0.06 |
ENSDART00000133057
ENSDART00000111129 |
ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr5_+_24569070 | 0.05 |
ENSDART00000012268
|
mrpl41
|
mitochondrial ribosomal protein L41 |
| chr16_+_55121654 | 0.05 |
ENSDART00000149449
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr12_-_3275183 | 0.05 |
ENSDART00000177055
|
CABZ01063170.1
|
ENSDARG00000107441 |
| chr24_+_28571936 | 0.05 |
ENSDART00000153933
|
CR933730.1
|
ENSDARG00000097841 |
| chr25_-_21718782 | 0.05 |
ENSDART00000175132
|
ENSDARG00000051873
|
ENSDARG00000051873 |
| chr6_+_43236957 | 0.05 |
ENSDART00000112474
|
arl6ip5a
|
ADP-ribosylation factor-like 6 interacting protein 5a |
| chr11_+_43127567 | 0.05 |
ENSDART00000172982
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr5_-_21548930 | 0.05 |
ENSDART00000002938
|
mtmr8
|
myotubularin related protein 8 |
| chr18_+_27753985 | 0.05 |
ENSDART00000141835
|
tspan18b
|
tetraspanin 18b |
| KN150171v1_+_3909 | 0.05 |
ENSDART00000171048
|
CABZ01071639.1
|
ENSDARG00000099435 |
| chr23_-_35384196 | 0.05 |
ENSDART00000138660
|
fbxo25
|
F-box protein 25 |
| chr2_+_41973443 | 0.05 |
|
|
|
| chr22_+_17484541 | 0.05 |
ENSDART00000088419
|
march2
|
membrane-associated ring finger (C3HC4) 2 |
| chr2_-_43997672 | 0.04 |
ENSDART00000005449
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr10_+_32739166 | 0.04 |
ENSDART00000063551
|
ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr25_+_6059393 | 0.04 |
ENSDART00000154658
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
| chr25_+_7858886 | 0.04 |
ENSDART00000171904
|
ucmab
|
upper zone of growth plate and cartilage matrix associated b |
| chr11_+_38858351 | 0.04 |
ENSDART00000155746
|
cdc42
|
cell division cycle 42 |
| chr1_-_57931565 | 0.04 |
ENSDART00000109528
|
adgre5b.3
|
adhesion G protein-coupled receptor E5b, duplicate 3 |
| chr2_+_33206349 | 0.04 |
ENSDART00000145588
|
rnf220a
|
ring finger protein 220a |
| chr16_-_52847642 | 0.04 |
ENSDART00000147236
|
azin1a
|
antizyme inhibitor 1a |
| chr14_-_33704021 | 0.04 |
ENSDART00000149396
ENSDART00000123607 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr6_-_16267327 | 0.04 |
ENSDART00000089445
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr22_-_19077300 | 0.04 |
ENSDART00000166295
|
polrmt
|
polymerase (RNA) mitochondrial (DNA directed) |
| chr22_+_39067994 | 0.04 |
|
|
|
| chr17_+_15520774 | 0.04 |
|
|
|
| chr10_-_2686054 | 0.04 |
ENSDART00000123754
|
mier3a
|
mesoderm induction early response 1, family member 3 a |
| chr10_+_27090911 | 0.04 |
ENSDART00000089095
|
wdr74
|
WD repeat domain 74 |
| chr25_-_6263829 | 0.04 |
ENSDART00000166084
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr10_+_19059663 | 0.04 |
ENSDART00000038674
|
tmem230a
|
transmembrane protein 230a |
| chr14_-_36057494 | 0.04 |
ENSDART00000052562
|
spata4
|
spermatogenesis associated 4 |
| chr1_+_58365570 | 0.04 |
ENSDART00000152669
|
gpr108
|
G protein-coupled receptor 108 |
| chr13_-_31246639 | 0.03 |
ENSDART00000026692
|
ubtd1a
|
ubiquitin domain containing 1a |
| chr4_+_26047568 | 0.03 |
ENSDART00000126474
|
si:ch211-265o23.1
|
si:ch211-265o23.1 |
| chr2_+_44692913 | 0.03 |
ENSDART00000155362
|
ENSDARG00000079124
|
ENSDARG00000079124 |
| chr9_+_2472749 | 0.03 |
ENSDART00000147034
|
gpr155a
|
G protein-coupled receptor 155a |
| chr21_-_22435721 | 0.03 |
|
|
|
| chr20_+_6600184 | 0.03 |
ENSDART00000136567
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr22_-_22696091 | 0.03 |
|
|
|
| chr8_-_25678085 | 0.03 |
ENSDART00000143938
|
sema3ga
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ga |
| chr1_+_11661380 | 0.03 |
ENSDART00000144475
|
clta
|
clathrin, light chain A |
| chr9_-_3853804 | 0.03 |
|
|
|
| chr13_-_12898263 | 0.03 |
ENSDART00000009499
|
whsc1
|
Wolf-Hirschhorn syndrome candidate 1 |
| chr13_-_24778352 | 0.03 |
|
|
|
| chr25_+_17308788 | 0.03 |
ENSDART00000164349
|
e2f4
|
E2F transcription factor 4 |
| chr21_-_22510751 | 0.03 |
ENSDART00000169870
|
myo5b
|
myosin VB |
| chr15_-_14162320 | 0.03 |
ENSDART00000156947
|
si:ch211-116o3.5
|
si:ch211-116o3.5 |
| chr8_-_31597206 | 0.03 |
ENSDART00000018886
|
ghra
|
growth hormone receptor a |
| chr24_-_13205000 | 0.03 |
ENSDART00000134482
|
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr9_+_17421457 | 0.03 |
ENSDART00000006256
|
zgc:101559
|
zgc:101559 |
| chr25_+_18467034 | 0.03 |
ENSDART00000170841
|
cav1
|
caveolin 1 |
| chr11_-_33355651 | 0.03 |
ENSDART00000033980
|
lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr4_+_11484516 | 0.02 |
ENSDART00000140954
|
asb13a.2
|
ankyrin repeat and SOCS box containing 13a, tandem duplicate 2 |
| chr6_-_12079553 | 0.02 |
ENSDART00000157058
|
si:dkey-276j7.1
|
si:dkey-276j7.1 |
| chr11_-_43125936 | 0.02 |
|
|
|
| chr19_-_7913229 | 0.02 |
ENSDART00000104703
|
znf687b
|
zinc finger protein 687b |
| chr9_-_3700395 | 0.02 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr9_+_26029780 | 0.02 |
ENSDART00000127135
|
gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr25_+_34628342 | 0.02 |
ENSDART00000155501
|
zgc:110434
|
zgc:110434 |
| chr2_+_10265922 | 0.02 |
ENSDART00000143876
|
cmpk
|
cytidylate kinase |
| chr13_+_24132494 | 0.02 |
ENSDART00000135992
|
abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr21_-_29995378 | 0.02 |
ENSDART00000178201
|
pwwp2a
|
PWWP domain containing 2A |
| chr3_-_27159896 | 0.02 |
ENSDART00000151742
|
BX908803.1
|
ENSDARG00000096260 |
| chr13_-_4005533 | 0.02 |
ENSDART00000132354
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr18_+_38928044 | 0.02 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 | 0.6 | GO:0002706 | regulation of lymphocyte mediated immunity(GO:0002706) |
| 0.2 | 0.6 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.1 | 0.3 | GO:0008206 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.1 | 0.3 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.1 | 0.4 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.4 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.0 | 0.3 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.0 | 0.1 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.1 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.2 | GO:0060416 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0006404 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.0 | 0.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) positive regulation of lipid transport(GO:0032370) |
| 0.0 | 0.1 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.1 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.0 | 0.5 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.5 | GO:0046463 | triglyceride biosynthetic process(GO:0019432) neutral lipid biosynthetic process(GO:0046460) acylglycerol biosynthetic process(GO:0046463) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.0 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.0 | 0.1 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.1 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.2 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.0 | GO:0099631 | postsynaptic endocytic zone cytoplasmic component(GO:0099631) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.5 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.7 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.1 | 0.4 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.2 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.0 | 0.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.8 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.3 | GO:0044325 | ion channel binding(GO:0044325) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |