Project
DANIO-CODE
Navigation
Downloads
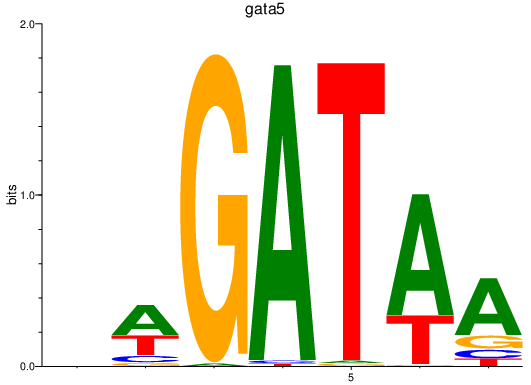
Results for gata5
Z-value: 0.51
Transcription factors associated with gata5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gata5
|
ENSDARG00000017821 | GATA binding protein 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gata5 | dr10_dc_chr23_+_7445760_7445883 | 0.61 | 1.2e-02 | Click! |
Activity profile of gata5 motif
Sorted Z-values of gata5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of gata5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_12613413 | 1.14 |
ENSDART00000166071
|
cyp2k16
|
cytochrome P450, family 2, subfamily K, polypeptide16 |
| chr3_+_31490043 | 1.05 |
ENSDART00000076636
|
fzd2
|
frizzled class receptor 2 |
| chr16_+_24069711 | 1.05 |
ENSDART00000142869
|
apoc2
|
apolipoprotein C-II |
| chr15_-_18425091 | 1.00 |
ENSDART00000155866
|
zbtb16b
|
zinc finger and BTB domain containing 16b |
| chr16_+_53238110 | 0.92 |
ENSDART00000102170
|
CABZ01053976.1
|
ENSDARG00000069929 |
| chr18_+_24932972 | 0.80 |
ENSDART00000008638
|
rgma
|
repulsive guidance molecule family member a |
| chr2_+_27739201 | 0.78 |
ENSDART00000145909
|
sepp1b
|
selenoprotein P, plasma, 1b |
| chr23_+_7445760 | 0.75 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr7_+_25649559 | 0.72 |
ENSDART00000026295
|
arrb2b
|
arrestin, beta 2b |
| chr6_-_8126955 | 0.72 |
ENSDART00000132412
|
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr6_+_43236843 | 0.67 |
ENSDART00000112474
|
arl6ip5a
|
ADP-ribosylation factor-like 6 interacting protein 5a |
| chr25_-_21692269 | 0.64 |
|
|
|
| chr7_-_37350985 | 0.63 |
ENSDART00000173523
|
nkd1
|
naked cuticle homolog 1 (Drosophila) |
| chr23_+_38239701 | 0.62 |
ENSDART00000148188
|
zgc:112994
|
zgc:112994 |
| chr10_-_17527124 | 0.61 |
ENSDART00000137905
|
nt5c2l1
|
5'-nucleotidase, cytosolic II, like 1 |
| chr7_+_24770873 | 0.61 |
ENSDART00000165314
|
AL954715.1
|
ENSDARG00000103682 |
| chr23_-_26150495 | 0.59 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr25_-_13631707 | 0.59 |
ENSDART00000169865
|
lcat
|
lecithin-cholesterol acyltransferase |
| chr3_-_39287733 | 0.59 |
|
|
|
| chr21_-_30074456 | 0.56 |
ENSDART00000014223
|
slc23a1
|
solute carrier family 23 (ascorbic acid transporter), member 1 |
| chr23_+_21610798 | 0.55 |
ENSDART00000111966
|
arhgef10lb
|
Rho guanine nucleotide exchange factor (GEF) 10-like b |
| chr5_+_27897765 | 0.52 |
ENSDART00000166737
ENSDART00000164418 |
nfr
|
notochord formation related |
| chr16_-_17439735 | 0.51 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr21_-_40811269 | 0.50 |
ENSDART00000148513
|
limk1b
|
LIM domain kinase 1b |
| chr11_+_27970922 | 0.47 |
ENSDART00000169360
ENSDART00000043756 |
ephb2b
|
eph receptor B2b |
| chr23_+_19018062 | 0.47 |
ENSDART00000104487
|
cox4i2
|
cytochrome c oxidase subunit IV isoform 2 |
| chr22_+_1568413 | 0.46 |
ENSDART00000175704
|
si:ch211-255f4.11
|
si:ch211-255f4.11 |
| chr3_+_32360862 | 0.44 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| KN150178v1_-_23644 | 0.43 |
ENSDART00000165583
|
soul3
|
heme-binding protein soul3 |
| chr9_+_32267615 | 0.42 |
|
|
|
| chr4_+_13453879 | 0.42 |
ENSDART00000145069
|
dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr7_+_39139117 | 0.40 |
ENSDART00000165594
|
CT030188.1
|
ENSDARG00000099385 |
| chr7_+_25793777 | 0.40 |
ENSDART00000174010
ENSDART00000173815 |
CT027989.1
|
ENSDARG00000105638 |
| chr2_+_27739142 | 0.39 |
ENSDART00000115071
|
sepp1b
|
selenoprotein P, plasma, 1b |
| chr20_-_47932043 | 0.38 |
ENSDART00000085624
|
ipo13
|
importin 13 |
| chr13_+_51154848 | 0.37 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr12_-_36138709 | 0.36 |
ENSDART00000130985
|
st6galnac
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase |
| chr15_-_11257445 | 0.36 |
|
|
|
| chr7_+_39139248 | 0.35 |
ENSDART00000165594
|
CT030188.1
|
ENSDARG00000099385 |
| chr2_+_4534596 | 0.33 |
ENSDART00000157744
ENSDART00000163111 ENSDART00000163986 |
wacb
|
WW domain containing adaptor with coiled-coil b |
| chr17_-_44133970 | 0.33 |
ENSDART00000126097
|
otx2
|
orthodenticle homeobox 2 |
| chr6_-_37765778 | 0.32 |
ENSDART00000149068
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr7_+_25794014 | 0.32 |
ENSDART00000174010
ENSDART00000173815 |
CT027989.1
|
ENSDARG00000105638 |
| chr19_-_41933808 | 0.32 |
ENSDART00000062080
|
chrac1
|
chromatin accessibility complex 1 |
| chr18_+_25016753 | 0.31 |
ENSDART00000099476
|
fam174b
|
family with sequence similarity 174, member B |
| chr23_-_1008307 | 0.30 |
ENSDART00000110588
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr22_-_21151096 | 0.30 |
ENSDART00000131743
|
si:ch211-87h14.3
|
si:ch211-87h14.3 |
| chr10_-_24422496 | 0.29 |
ENSDART00000141332
ENSDART00000100772 |
slc43a2b
|
solute carrier family 43 (amino acid system L transporter), member 2b |
| chr19_-_11096996 | 0.27 |
ENSDART00000010997
|
tpm3
|
tropomyosin 3 |
| chr11_+_11809234 | 0.27 |
ENSDART00000168346
|
CR848705.1
|
ENSDARG00000102054 |
| chr8_-_31597252 | 0.26 |
ENSDART00000018886
|
ghra
|
growth hormone receptor a |
| chr15_+_25700036 | 0.26 |
ENSDART00000135409
ENSDART00000162240 |
tsr1
|
TSR1, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr11_+_38858351 | 0.25 |
ENSDART00000155746
|
cdc42
|
cell division cycle 42 |
| chr9_+_32267297 | 0.24 |
|
|
|
| chr2_-_53797503 | 0.23 |
ENSDART00000057053
|
im:7138239
|
im:7138239 |
| chr9_-_6683651 | 0.23 |
ENSDART00000061593
|
pou3f3a
|
POU class 3 homeobox 3a |
| chr4_+_22749707 | 0.22 |
|
|
|
| chr6_-_54103765 | 0.21 |
ENSDART00000083880
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr6_+_19167652 | 0.21 |
ENSDART00000086619
|
prkca
|
protein kinase C, alpha |
| chr2_-_53248164 | 0.19 |
ENSDART00000114682
|
ralbp1
|
ralA binding protein 1 |
| chr5_+_18421698 | 0.19 |
ENSDART00000051621
|
pgam5
|
PGAM family member 5, serine/threonine protein phosphatase, mitochondrial |
| chr9_+_50670635 | 0.18 |
ENSDART00000179475
|
CABZ01083707.1
|
ENSDARG00000107278 |
| chr10_+_42761097 | 0.18 |
ENSDART00000075259
|
fgfr1b
|
fibroblast growth factor receptor 1b |
| chr2_-_53797678 | 0.17 |
ENSDART00000057053
|
im:7138239
|
im:7138239 |
| chr3_-_7252474 | 0.16 |
|
|
|
| chr19_-_4769990 | 0.16 |
|
|
|
| chr19_+_341775 | 0.15 |
ENSDART00000151013
|
ensaa
|
endosulfine alpha a |
| chr6_+_24561921 | 0.15 |
ENSDART00000169283
|
znf644b
|
zinc finger protein 644b |
| chr17_-_17744885 | 0.14 |
ENSDART00000155261
|
slirp
|
SRA stem-loop interacting RNA binding protein |
| chr8_-_25678416 | 0.13 |
ENSDART00000143938
|
sema3ga
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ga |
| chr16_+_12922721 | 0.13 |
ENSDART00000008535
|
u2af2a
|
U2 small nuclear RNA auxiliary factor 2a |
| chr16_-_30463239 | 0.12 |
ENSDART00000003752
|
cct3
|
chaperonin containing TCP1, subunit 3 (gamma) |
| chr3_-_14545180 | 0.12 |
ENSDART00000133850
|
gadd45gip1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr6_+_43236957 | 0.11 |
ENSDART00000112474
|
arl6ip5a
|
ADP-ribosylation factor-like 6 interacting protein 5a |
| chr15_+_22078226 | 0.11 |
ENSDART00000079504
|
ankk1
|
ankyrin repeat and kinase domain containing 1 |
| chr7_+_34394526 | 0.10 |
|
|
|
| chr13_-_4005533 | 0.10 |
ENSDART00000132354
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr3_-_13771322 | 0.10 |
ENSDART00000159177
ENSDART00000165174 |
si:dkey-61n16.5
|
si:dkey-61n16.5 |
| chr8_+_8934762 | 0.10 |
ENSDART00000133137
|
bcap31
|
B-cell receptor-associated protein 31 |
| chr3_+_33309011 | 0.09 |
ENSDART00000164322
|
gtpbp1
|
GTP binding protein 1 |
| chr6_-_54103833 | 0.09 |
ENSDART00000083880
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr11_+_5845373 | 0.09 |
ENSDART00000111374
|
ndufs7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, (NADH-coenzyme Q reductase) |
| chr1_+_45147775 | 0.09 |
ENSDART00000179047
|
map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr4_+_13453833 | 0.09 |
ENSDART00000145069
|
dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr20_+_23602537 | 0.08 |
|
|
|
| chr19_-_11096542 | 0.08 |
|
|
|
| chr16_+_24818799 | 0.08 |
ENSDART00000155217
|
si:dkey-79d12.4
|
si:dkey-79d12.4 |
| chr19_-_7913229 | 0.06 |
ENSDART00000104703
|
znf687b
|
zinc finger protein 687b |
| chr4_-_5904787 | 0.06 |
ENSDART00000169439
|
arl1
|
ADP-ribosylation factor-like 1 |
| chr20_-_35173500 | 0.06 |
ENSDART00000019196
|
lft1
|
lefty1 |
| chr8_+_6533379 | 0.06 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr24_-_7602759 | 0.06 |
ENSDART00000041714
|
atp6v0a1b
|
ATPase, H+ transporting, lysosomal V0 subunit a1b |
| chr22_+_35090105 | 0.05 |
|
|
|
| chr1_+_58365570 | 0.05 |
ENSDART00000152669
|
gpr108
|
G protein-coupled receptor 108 |
| chr20_-_10500595 | 0.05 |
ENSDART00000064112
|
glrx5
|
glutaredoxin 5 homolog (S. cerevisiae) |
| chr7_-_68989537 | 0.05 |
ENSDART00000176894
|
phlpp2
|
PH domain and leucine rich repeat protein phosphatase 2 |
| chr6_-_16267366 | 0.04 |
ENSDART00000089445
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr14_+_6655541 | 0.04 |
ENSDART00000148447
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr16_-_28333633 | 0.04 |
ENSDART00000121671
|
si:dkey-12j5.1
|
si:dkey-12j5.1 |
| chr3_-_40791035 | 0.04 |
ENSDART00000055201
|
foxk1
|
forkhead box K1 |
| chr22_-_22696091 | 0.04 |
|
|
|
| chr23_-_19055201 | 0.03 |
ENSDART00000147617
|
bcl2l1
|
bcl2-like 1 |
| chr8_-_25678085 | 0.03 |
ENSDART00000143938
|
sema3ga
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ga |
| chr11_+_14373731 | 0.02 |
ENSDART00000161879
|
BX571942.1
|
ENSDARG00000099220 |
| chr10_+_21699252 | 0.02 |
ENSDART00000160464
|
pcdh1gb2
|
protocadherin 1 gamma b 2 |
| chr23_+_29431388 | 0.02 |
ENSDART00000027255
|
tardbpl
|
TAR DNA binding protein, like |
| chr10_+_11439845 | 0.02 |
ENSDART00000064210
|
cwc27
|
CWC27 spliceosome-associated protein homolog (S. cerevisiae) |
| chr19_+_40792612 | 0.02 |
ENSDART00000017917
|
vps50
|
VPS50 EARP/GARPII complex subunit |
| chr5_+_17543117 | 0.01 |
ENSDART00000132164
|
hira
|
histone cell cycle regulator a |
| chr12_-_16035939 | 0.01 |
ENSDART00000090881
|
si:dkey-100n10.2
|
si:dkey-100n10.2 |
| chr18_-_20880226 | 0.00 |
ENSDART00000090079
|
synm
|
synemin, intermediate filament protein |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.2 | 1.0 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.2 | 0.7 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) regulation of protein binding(GO:0043393) |
| 0.2 | 0.7 | GO:0048618 | post-embryonic morphogenesis(GO:0009886) post-embryonic foregut morphogenesis(GO:0048618) |
| 0.2 | 0.6 | GO:1903533 | regulation of protein targeting(GO:1903533) |
| 0.1 | 0.4 | GO:0002706 | regulation of lymphocyte mediated immunity(GO:0002706) |
| 0.1 | 0.6 | GO:0015882 | L-ascorbic acid transport(GO:0015882) |
| 0.1 | 0.7 | GO:0045453 | bone resorption(GO:0045453) |
| 0.1 | 1.1 | GO:0007223 | Wnt signaling pathway involved in heart development(GO:0003306) Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.1 | 1.1 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 1.0 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 1.1 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.0 | 0.2 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.4 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.5 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.7 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.3 | GO:0000479 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.8 | GO:0051051 | negative regulation of transport(GO:0051051) |
| 0.0 | 0.3 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.2 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 0.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.3 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.7 | GO:0003993 | acid phosphatase activity(GO:0003993) ferric iron binding(GO:0008199) |
| 0.1 | 0.8 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.2 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.1 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.3 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 1.1 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.4 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.4 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |