Project
DANIO-CODE
Navigation
Downloads
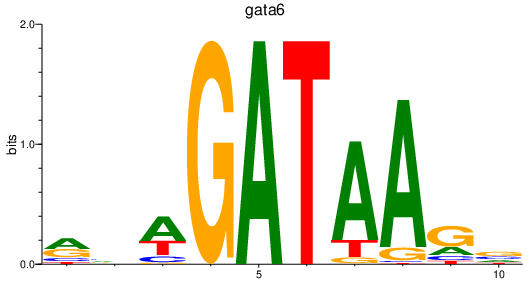
Results for gata6
Z-value: 0.93
Transcription factors associated with gata6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gata6
|
ENSDARG00000103589 | GATA binding protein 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gata6 | dr10_dc_chr2_+_4366169_4366297 | 0.76 | 6.6e-04 | Click! |
Activity profile of gata6 motif
Sorted Z-values of gata6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of gata6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_32160464 | 2.74 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr10_+_10843384 | 2.73 |
ENSDART00000130283
|
ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr25_+_21732255 | 2.53 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr12_-_20228689 | 2.44 |
ENSDART00000066384
|
hbbe2
|
hemoglobin beta embryonic-2 |
| chr10_-_13814731 | 1.92 |
ENSDART00000145103
|
cntfr
|
ciliary neurotrophic factor receptor |
| chr23_+_7445760 | 1.89 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr8_+_7740132 | 1.86 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr17_+_19479310 | 1.79 |
ENSDART00000077804
|
slc22a15
|
solute carrier family 22, member 15 |
| chr13_+_2774422 | 1.61 |
ENSDART00000162362
|
wu:fj16a03
|
wu:fj16a03 |
| chr10_-_32614936 | 1.54 |
ENSDART00000143301
|
mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr5_-_63487161 | 1.44 |
ENSDART00000048395
|
cmlc1
|
cardiac myosin light chain-1 |
| chr23_-_10747605 | 1.39 |
|
|
|
| chr2_+_21342233 | 1.37 |
ENSDART00000062563
|
rreb1b
|
ras responsive element binding protein 1b |
| chr13_-_22901327 | 1.36 |
ENSDART00000056523
|
hkdc1
|
hexokinase domain containing 1 |
| chr16_-_17439735 | 1.34 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr23_+_25782195 | 1.30 |
ENSDART00000060059
|
rbms2b
|
RNA binding motif, single stranded interacting protein 2b |
| chr22_+_1568413 | 1.30 |
ENSDART00000175704
|
si:ch211-255f4.11
|
si:ch211-255f4.11 |
| chr8_-_40287789 | 1.29 |
ENSDART00000074125
|
aplnra
|
apelin receptor a |
| chr20_+_15653121 | 1.21 |
ENSDART00000152702
|
jun
|
jun proto-oncogene |
| chr16_+_21090083 | 1.17 |
ENSDART00000052662
|
hoxa13b
|
homeobox A13b |
| chr20_+_40247918 | 1.15 |
ENSDART00000121818
|
trdn
|
triadin |
| chr24_-_39970858 | 1.13 |
ENSDART00000134251
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
| chr6_+_58921655 | 1.13 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr5_-_37521979 | 1.10 |
ENSDART00000141791
ENSDART00000170528 |
si:ch211-284e13.6
|
si:ch211-284e13.6 |
| chr7_-_73530653 | 1.10 |
ENSDART00000166633
ENSDART00000009888 ENSDART00000171254 |
casq1b
|
calsequestrin 1b |
| chr6_-_12079553 | 1.09 |
ENSDART00000157058
|
si:dkey-276j7.1
|
si:dkey-276j7.1 |
| chr20_+_6600184 | 1.05 |
ENSDART00000136567
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr8_+_23464087 | 1.05 |
ENSDART00000026316
|
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr16_+_13534390 | 1.04 |
ENSDART00000157396
|
ENSDARG00000022807
|
ENSDARG00000022807 |
| chr1_+_1657493 | 1.01 |
ENSDART00000103850
|
atp1a1a.3
|
ATPase, Na+/K+ transporting, alpha 1a polypeptide, tandem duplicate 3 |
| chr8_-_18502159 | 0.98 |
ENSDART00000148802
ENSDART00000149081 ENSDART00000148962 |
nexn
|
nexilin (F actin binding protein) |
| chr5_+_49103778 | 0.98 |
ENSDART00000155405
|
CR751602.2
|
ENSDARG00000098045 |
| chr19_-_27730419 | 0.98 |
ENSDART00000151047
|
sh3bp5lb
|
SH3-binding domain protein 5-like, b |
| chr19_+_4996328 | 0.97 |
ENSDART00000165082
|
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr14_+_33382973 | 0.95 |
ENSDART00000132488
|
apln
|
apelin |
| chr3_+_52901602 | 0.94 |
ENSDART00000114343
|
brd4
|
bromodomain containing 4 |
| chr5_+_34022151 | 0.94 |
ENSDART00000141338
|
enc1
|
ectodermal-neural cortex 1 |
| chr17_-_13072982 | 0.92 |
ENSDART00000178859
|
CABZ01047998.1
|
ENSDARG00000105945 |
| chr3_-_56969897 | 0.91 |
|
|
|
| chr20_+_6600246 | 0.90 |
ENSDART00000136567
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr13_+_24132494 | 0.90 |
ENSDART00000135992
|
abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr20_+_23339698 | 0.87 |
ENSDART00000074167
|
ociad2
|
OCIA domain containing 2 |
| chr23_-_30114528 | 0.86 |
ENSDART00000131209
|
ccdc187
|
coiled-coil domain containing 187 |
| chr6_-_37765778 | 0.80 |
ENSDART00000149068
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr8_-_17580655 | 0.77 |
|
|
|
| chr6_-_54103765 | 0.75 |
ENSDART00000083880
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr25_+_18467034 | 0.75 |
ENSDART00000170841
|
cav1
|
caveolin 1 |
| chr16_+_26576242 | 0.74 |
ENSDART00000039746
|
epb41b
|
erythrocyte membrane protein band 4.1b |
| chr16_-_5936637 | 0.73 |
ENSDART00000085678
|
trak1
|
trafficking protein, kinesin binding 1 |
| chr22_-_22696091 | 0.70 |
|
|
|
| chr11_-_1374026 | 0.69 |
ENSDART00000172953
|
rpl29
|
ribosomal protein L29 |
| chr3_-_26073676 | 0.69 |
ENSDART00000169123
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr14_-_33704021 | 0.68 |
ENSDART00000149396
ENSDART00000123607 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr23_-_23474703 | 0.65 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr12_-_26339002 | 0.65 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr11_-_9479592 | 0.65 |
ENSDART00000175829
|
CU179643.1
|
ENSDARG00000086489 |
| chr10_-_13814679 | 0.64 |
ENSDART00000145103
|
cntfr
|
ciliary neurotrophic factor receptor |
| chr25_-_35698109 | 0.63 |
ENSDART00000134928
|
nfatc3b
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3b |
| chr21_+_25899113 | 0.62 |
ENSDART00000141149
|
caln2
|
calneuron 2 |
| chr11_-_27253835 | 0.61 |
ENSDART00000065889
|
wnt7aa
|
wingless-type MMTV integration site family, member 7Aa |
| chr17_-_24666272 | 0.61 |
ENSDART00000105457
|
morn2
|
MORN repeat containing 2 |
| chr8_-_6833841 | 0.60 |
ENSDART00000005321
|
neflb
|
neurofilament, light polypeptide b |
| KN150171v1_+_3909 | 0.59 |
ENSDART00000171048
|
CABZ01071639.1
|
ENSDARG00000099435 |
| chr23_+_25930072 | 0.59 |
ENSDART00000124103
|
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr16_-_22860052 | 0.58 |
ENSDART00000127570
|
pbxip1b
|
pre-B-cell leukemia homeobox interacting protein 1b |
| chr3_-_26073923 | 0.58 |
ENSDART00000103748
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr14_+_33382672 | 0.56 |
ENSDART00000075312
|
apln
|
apelin |
| chr8_+_6533379 | 0.56 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr16_-_13652300 | 0.52 |
ENSDART00000144140
|
BX323458.2
|
ENSDARG00000094614 |
| chr3_-_30810423 | 0.52 |
ENSDART00000076830
|
coro1a
|
coronin, actin binding protein, 1A |
| chr12_+_33879261 | 0.51 |
|
|
|
| chr11_-_35313757 | 0.51 |
ENSDART00000031441
|
sema3fb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fb |
| chr14_-_31918174 | 0.50 |
ENSDART00000052949
|
fgf13a
|
fibroblast growth factor 13a |
| chr14_-_47909935 | 0.49 |
ENSDART00000058789
|
qdpra
|
quinoid dihydropteridine reductase a |
| chr3_+_20007257 | 0.48 |
ENSDART00000125281
|
ngfra
|
nerve growth factor receptor a (TNFR superfamily, member 16) |
| chr21_-_25719352 | 0.47 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr11_-_9479689 | 0.42 |
ENSDART00000175829
|
CU179643.1
|
ENSDARG00000086489 |
| chr10_-_25629613 | 0.42 |
ENSDART00000131640
|
tiam1a
|
T-cell lymphoma invasion and metastasis 1a |
| chr2_-_52202004 | 0.38 |
ENSDART00000165350
|
BX908782.1
|
ENSDARG00000098957 |
| chr8_+_25594106 | 0.37 |
|
|
|
| chr19_+_38270817 | 0.33 |
|
|
|
| chr9_-_9250896 | 0.30 |
ENSDART00000121665
|
cbsb
|
cystathionine-beta-synthase b |
| chr13_+_24132236 | 0.30 |
ENSDART00000088005
|
abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr3_+_28371725 | 0.29 |
ENSDART00000151081
|
sept12
|
septin 12 |
| chr25_-_22955066 | 0.29 |
ENSDART00000024633
|
dusp8a
|
dual specificity phosphatase 8a |
| chr20_+_40248154 | 0.29 |
ENSDART00000121818
|
trdn
|
triadin |
| chr6_+_43236843 | 0.27 |
ENSDART00000112474
|
arl6ip5a
|
ADP-ribosylation factor-like 6 interacting protein 5a |
| chr15_-_19162052 | 0.26 |
|
|
|
| chr6_-_43782889 | 0.25 |
|
|
|
| chr15_+_31956131 | 0.23 |
|
|
|
| chr23_+_20936600 | 0.23 |
ENSDART00000129992
|
pax7b
|
paired box 7b |
| chr14_+_34154895 | 0.23 |
ENSDART00000147756
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr7_+_26274252 | 0.21 |
ENSDART00000164824
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr17_-_29885237 | 0.20 |
ENSDART00000009104
|
esrrga
|
estrogen-related receptor gamma a |
| chr4_+_16736949 | 0.20 |
ENSDART00000161394
|
tcp11l2
|
t-complex 11, testis-specific-like 2 |
| chr19_-_7913229 | 0.19 |
ENSDART00000104703
|
znf687b
|
zinc finger protein 687b |
| chr17_+_27384572 | 0.19 |
|
|
|
| chr7_-_22519196 | 0.18 |
ENSDART00000129919
|
ENSDARG00000091111
|
ENSDARG00000091111 |
| chr10_-_24422496 | 0.17 |
ENSDART00000141332
ENSDART00000100772 |
slc43a2b
|
solute carrier family 43 (amino acid system L transporter), member 2b |
| chr20_-_16022816 | 0.17 |
ENSDART00000152828
|
fam20b
|
family with sequence similarity 20, member B (H. sapiens) |
| chr6_-_42379717 | 0.16 |
ENSDART00000133647
|
emc3
|
ER membrane protein complex subunit 3 |
| chr1_-_499082 | 0.16 |
ENSDART00000144406
ENSDART00000031635 |
ercc5
|
excision repair cross-complementation group 5 |
| chr21_+_39055276 | 0.15 |
ENSDART00000075958
|
slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr6_-_54103833 | 0.13 |
ENSDART00000083880
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr17_+_1660922 | 0.11 |
ENSDART00000159190
|
srp14
|
signal recognition particle 14 |
| chr14_+_6655894 | 0.11 |
ENSDART00000148394
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr17_+_8018808 | 0.11 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr13_+_18235298 | 0.10 |
ENSDART00000133057
ENSDART00000111129 |
ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr8_+_7740024 | 0.08 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr18_+_38928044 | 0.08 |
|
|
|
| chr6_-_58921392 | 0.08 |
|
|
|
| chr19_-_3208863 | 0.06 |
|
|
|
| chr24_+_3447129 | 0.04 |
ENSDART00000134598
|
wdr37
|
WD repeat domain 37 |
| chr13_-_22901265 | 0.04 |
ENSDART00000056523
|
hkdc1
|
hexokinase domain containing 1 |
| chr14_+_6656015 | 0.03 |
ENSDART00000148394
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr8_+_7739964 | 0.03 |
ENSDART00000171325
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr1_+_58365570 | 0.01 |
ENSDART00000152669
|
gpr108
|
G protein-coupled receptor 108 |
| chr13_+_1665226 | 0.01 |
ENSDART00000029343
|
col21a1
|
collagen, type XXI, alpha 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.5 | 1.5 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.5 | 1.9 | GO:0009886 | post-embryonic morphogenesis(GO:0009886) post-embryonic foregut morphogenesis(GO:0048618) |
| 0.4 | 1.1 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.4 | 1.1 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.4 | 2.6 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.3 | 2.0 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.3 | 2.5 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.2 | 1.0 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.2 | 0.9 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.2 | 2.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 2.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 1.4 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.2 | 0.7 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.2 | 0.9 | GO:1904323 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) positive regulation of calcium ion transport into cytosol(GO:0010524) positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904323) positive regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904325) |
| 0.2 | 0.9 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.3 | GO:0033238 | regulation of cellular amino acid metabolic process(GO:0006521) regulation of cellular amine metabolic process(GO:0033238) regulation of sulfur metabolic process(GO:0042762) |
| 0.1 | 0.5 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 0.6 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 1.0 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 0.7 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 2.0 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.1 | 0.2 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.1 | 0.2 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.1 | 0.6 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.6 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.6 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.7 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.5 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.3 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.2 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.7 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 0.6 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 0.7 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.6 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.4 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 1.2 | GO:0033333 | fin development(GO:0033333) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | GO:0031838 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 1.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.6 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 1.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.2 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.0 | 1.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 3.2 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.7 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.3 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.3 | 2.5 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.3 | 0.9 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.3 | 1.5 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.2 | 0.5 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 0.9 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 1.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 1.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.6 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.5 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 0.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.0 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 1.5 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.1 | 2.6 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.1 | 1.0 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 2.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 2.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 1.0 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 0.7 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 1.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 2.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 1.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.9 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |