Project
DANIO-CODE
Navigation
Downloads
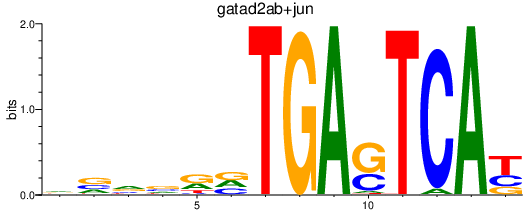
Results for gatad2ab+jun_fosl2_smarcc1b
Z-value: 0.66
Transcription factors associated with gatad2ab+jun_fosl2_smarcc1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gatad2ab
|
ENSDARG00000006192 | GATA zinc finger domain containing 2Ab |
|
jun
|
ENSDARG00000043531 | Jun proto-oncogene, AP-1 transcription factor subunit |
|
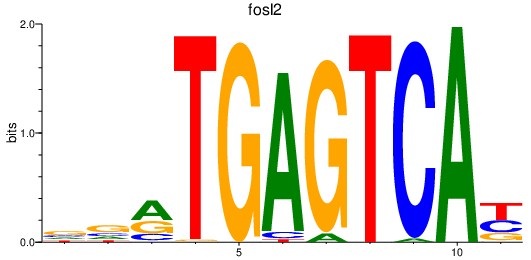
fosl2
|
ENSDARG00000040623 | fos-like antigen 2 |
|
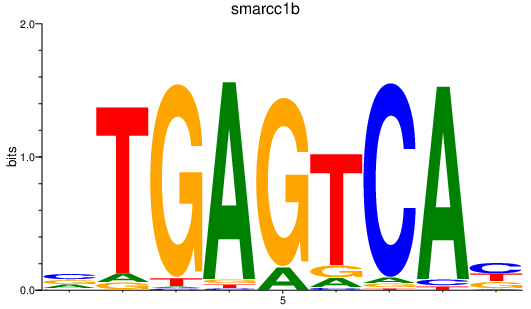
smarcc1b
|
ENSDARG00000098919 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| jun | dr10_dc_chr20_+_15653121_15653193 | 0.66 | 5.0e-03 | Click! |
| smarcc1b | dr10_dc_chr19_-_19806070_19806161 | -0.61 | 1.2e-02 | Click! |
| gatad2ab | dr10_dc_chr22_+_18294579_18294593 | -0.48 | 5.7e-02 | Click! |
| fosl2 | dr10_dc_chr17_+_41313762_41313801 | -0.13 | 6.2e-01 | Click! |
Activity profile of gatad2ab+jun_fosl2_smarcc1b motif
Sorted Z-values of gatad2ab+jun_fosl2_smarcc1b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of gatad2ab+jun_fosl2_smarcc1b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_7501777 | 2.35 |
ENSDART00000081741
|
rab25a
|
RAB25, member RAS oncogene family a |
| chr21_+_25729090 | 2.26 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr16_+_17808623 | 2.05 |
ENSDART00000149596
|
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr8_-_38168395 | 2.02 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr16_+_42925950 | 1.60 |
ENSDART00000159730
ENSDART00000014956 |
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr19_+_17451381 | 1.56 |
ENSDART00000167602
|
spaca4l
|
sperm acrosome associated 4 like |
| chr20_-_26632676 | 1.53 |
ENSDART00000131994
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr25_+_18487408 | 1.50 |
ENSDART00000148741
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr3_+_24067387 | 1.48 |
ENSDART00000055609
|
atf4b
|
activating transcription factor 4b |
| chr20_+_30948175 | 1.45 |
|
|
|
| chr23_+_25930072 | 1.44 |
ENSDART00000124103
|
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr8_+_25880895 | 1.43 |
ENSDART00000124300
|
rhoab
|
ras homolog gene family, member Ab |
| chr6_+_56157608 | 1.41 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr25_-_22089794 | 1.40 |
ENSDART00000139110
|
pkp3a
|
plakophilin 3a |
| chr5_-_30324182 | 1.39 |
ENSDART00000153909
|
spns2
|
spinster homolog 2 (Drosophila) |
| chr6_-_22504772 | 1.30 |
ENSDART00000170039
|
sept9b
|
septin 9b |
| chr10_-_3294792 | 1.26 |
ENSDART00000109131
|
slc25a1b
|
slc25a1 solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1b |
| chr25_-_18234069 | 1.20 |
ENSDART00000104496
|
dusp6
|
dual specificity phosphatase 6 |
| chr19_-_7576069 | 1.18 |
ENSDART00000148836
|
rfx5
|
regulatory factor X, 5 |
| chr4_+_1750689 | 1.14 |
ENSDART00000146779
|
slc38a2
|
solute carrier family 38, member 2 |
| chr14_-_40454194 | 1.14 |
ENSDART00000166621
|
ef1
|
E74-like factor 1 (ets domain transcription factor) |
| chr14_-_2008649 | 1.11 |
ENSDART00000161817
|
pcdh2g16
|
protocadherin 2 gamma 16 |
| chr9_-_48673183 | 1.08 |
ENSDART00000140185
ENSDART00000134185 |
col28a2a
|
collagen, type XXVIII, alpha 2a |
| chr11_-_28367271 | 1.07 |
ENSDART00000065853
|
dhrs3b
|
dehydrogenase/reductase (SDR family) member 3b |
| chr10_-_20524670 | 1.07 |
|
|
|
| chr22_-_15567180 | 1.05 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr7_+_44373815 | 1.04 |
ENSDART00000170721
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr7_+_23306546 | 1.04 |
ENSDART00000049885
|
si:dkey-172j4.3
|
si:dkey-172j4.3 |
| chr13_-_36996246 | 1.01 |
ENSDART00000133242
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr6_+_56163589 | 0.98 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr4_-_6800721 | 0.97 |
ENSDART00000099467
|
ifrd1
|
interferon-related developmental regulator 1 |
| chr1_+_41148042 | 0.95 |
ENSDART00000145170
ENSDART00000136879 |
smox
|
spermine oxidase |
| chr5_+_23585492 | 0.94 |
ENSDART00000135083
|
tp53
|
tumor protein p53 |
| chr21_+_30757831 | 0.91 |
ENSDART00000139486
|
ENSDARG00000030006
|
ENSDARG00000030006 |
| chr14_-_17282615 | 0.89 |
ENSDART00000006716
ENSDART00000136242 |
selt2
|
selenoprotein T, 2 |
| chr7_-_26165200 | 0.89 |
ENSDART00000123395
|
her8a
|
hairy-related 8a |
| chr11_+_26371444 | 0.87 |
ENSDART00000042322
|
map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr5_-_65349550 | 0.85 |
ENSDART00000164228
|
nrarpb
|
notch-regulated ankyrin repeat protein b |
| chr17_+_25503946 | 0.85 |
|
|
|
| chr25_+_18487313 | 0.85 |
ENSDART00000148741
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr3_-_39346621 | 0.81 |
ENSDART00000135192
ENSDART00000013553 ENSDART00000167289 |
zgc:100868
|
zgc:100868 |
| chr22_+_37934447 | 0.80 |
ENSDART00000076082
|
fetub
|
fetuin B |
| chr10_+_4987494 | 0.79 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr3_+_57878398 | 0.79 |
ENSDART00000047418
|
notum1a
|
notum pectinacetylesterase homolog 1a (Drosophila) |
| chr1_-_51862897 | 0.79 |
ENSDART00000136469
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr18_+_45121016 | 0.78 |
ENSDART00000172328
|
gyltl1b
|
glycosyltransferase-like 1b |
| chr7_-_52848084 | 0.78 |
ENSDART00000172179
ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr23_-_30119058 | 0.77 |
ENSDART00000103480
|
ccdc187
|
coiled-coil domain containing 187 |
| chr20_+_28465622 | 0.77 |
ENSDART00000146905
ENSDART00000103355 |
rhov
|
ras homolog family member V |
| chr22_+_11726312 | 0.75 |
ENSDART00000155366
|
krt96
|
keratin 96 |
| KN149946v1_-_12861 | 0.74 |
ENSDART00000163016
|
CABZ01048402.2
|
ENSDARG00000103875 |
| chr15_-_19836573 | 0.74 |
ENSDART00000114888
|
picalmb
|
phosphatidylinositol binding clathrin assembly protein b |
| chr6_-_47928632 | 0.74 |
|
|
|
| chr17_+_28657998 | 0.74 |
ENSDART00000159067
|
hectd1
|
HECT domain containing 1 |
| chr11_+_37639045 | 0.73 |
ENSDART00000111157
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr3_-_25886553 | 0.73 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr16_+_55008251 | 0.72 |
ENSDART00000176995
|
CABZ01087388.1
|
ENSDARG00000106538 |
| chr1_-_416138 | 0.72 |
ENSDART00000092524
|
rasa3
|
RAS p21 protein activator 3 |
| chr23_-_1008307 | 0.71 |
ENSDART00000110588
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr3_+_26896869 | 0.70 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr16_-_31517906 | 0.70 |
ENSDART00000145691
|
ENSDARG00000054814
|
ENSDARG00000054814 |
| chr14_+_33542705 | 0.70 |
ENSDART00000019396
|
clic2
|
chloride intracellular channel 2 |
| chr1_+_52365033 | 0.69 |
ENSDART00000133411
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr13_-_3801122 | 0.68 |
|
|
|
| chr13_+_46487890 | 0.67 |
|
|
|
| chr5_+_41680352 | 0.67 |
ENSDART00000142855
|
tbx6l
|
T-box 6, like |
| chr6_-_47928548 | 0.66 |
|
|
|
| chr10_-_20524586 | 0.65 |
|
|
|
| chr8_+_49789789 | 0.65 |
ENSDART00000083790
|
ntrk2a
|
neurotrophic tyrosine kinase, receptor, type 2a |
| chr13_+_1051015 | 0.64 |
ENSDART00000033528
|
tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr10_-_6453139 | 0.63 |
ENSDART00000168549
|
ca9
|
carbonic anhydrase IX |
| chr11_-_28367400 | 0.62 |
ENSDART00000065853
|
dhrs3b
|
dehydrogenase/reductase (SDR family) member 3b |
| chr10_+_16111842 | 0.62 |
ENSDART00000141654
|
megf10
|
multiple EGF-like-domains 10 |
| chr13_+_35213326 | 0.61 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr14_-_26138828 | 0.60 |
ENSDART00000140173
|
si:dkeyp-110e4.6
|
si:dkeyp-110e4.6 |
| chr7_+_65532390 | 0.60 |
ENSDART00000156683
|
CT573494.2
|
ENSDARG00000097673 |
| chr5_+_36487425 | 0.59 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr8_+_30491476 | 0.59 |
ENSDART00000062073
|
ENSDARG00000042329
|
ENSDARG00000042329 |
| chr23_-_36805789 | 0.58 |
|
|
|
| chr1_-_5048203 | 0.57 |
ENSDART00000150863
ENSDART00000163417 |
nrp2a
|
neuropilin 2a |
| chr23_-_28367816 | 0.57 |
ENSDART00000003548
|
znf385a
|
zinc finger protein 385A |
| chr2_-_39710140 | 0.57 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr22_+_16471319 | 0.56 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr19_-_41819752 | 0.56 |
ENSDART00000167772
|
shfm1
|
split hand/foot malformation (ectrodactyly) type 1 |
| chr5_+_37184963 | 0.56 |
ENSDART00000053511
|
myo1ca
|
myosin Ic, paralog a |
| chr16_+_33702010 | 0.56 |
ENSDART00000143757
|
fhl3a
|
four and a half LIM domains 3a |
| chr3_+_28450576 | 0.55 |
ENSDART00000150893
|
sept12
|
septin 12 |
| chr6_+_29800606 | 0.55 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr15_-_12484651 | 0.55 |
ENSDART00000162258
|
BX901957.1
|
ENSDARG00000098363 |
| chr20_-_25726868 | 0.55 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr9_-_2588401 | 0.55 |
ENSDART00000161018
|
sp9
|
sp9 transcription factor |
| chr12_+_37005712 | 0.55 |
|
|
|
| chr16_+_24045774 | 0.55 |
ENSDART00000133484
|
apoeb
|
apolipoprotein Eb |
| chr9_-_2416300 | 0.54 |
|
|
|
| chr7_+_38455040 | 0.54 |
|
|
|
| chr20_+_29307039 | 0.53 |
ENSDART00000152949
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr20_-_32543497 | 0.53 |
ENSDART00000026635
|
nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr14_+_34146377 | 0.53 |
ENSDART00000131861
|
tmsb2
|
thymosin beta 2 |
| chr10_+_35535209 | 0.53 |
ENSDART00000109705
|
phldb2a
|
pleckstrin homology-like domain, family B, member 2a |
| chr19_-_19888074 | 0.53 |
|
|
|
| chr5_+_15319430 | 0.52 |
ENSDART00000162003
|
hspb8
|
heat shock protein b8 |
| chr18_+_21419735 | 0.52 |
ENSDART00000144523
|
necab2
|
N-terminal EF-hand calcium binding protein 2 |
| KN149817v1_+_2207 | 0.51 |
|
|
|
| chr15_-_18110169 | 0.51 |
|
|
|
| chr25_-_18044103 | 0.50 |
ENSDART00000113581
|
kitlga
|
kit ligand a |
| chr4_+_5308883 | 0.50 |
ENSDART00000150366
|
si:ch211-214j24.10
|
si:ch211-214j24.10 |
| chr1_-_24486146 | 0.49 |
ENSDART00000144711
|
tmem154
|
transmembrane protein 154 |
| chr20_+_34867305 | 0.49 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr9_-_12726136 | 0.49 |
|
|
|
| chr21_-_20983913 | 0.49 |
ENSDART00000132091
|
eif4ebp1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr15_+_23508351 | 0.48 |
ENSDART00000162997
|
phykpl
|
5-phosphohydroxy-L-lysine phospho-lyase |
| chr12_-_1931281 | 0.48 |
ENSDART00000005676
ENSDART00000127937 |
sox9a
|
SRY (sex determining region Y)-box 9a |
| chr6_+_42478185 | 0.48 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr3_+_37433008 | 0.48 |
ENSDART00000055225
|
wnt9b
|
wingless-type MMTV integration site family, member 9B |
| chr24_-_37407313 | 0.48 |
|
|
|
| chr8_-_15071283 | 0.47 |
|
|
|
| chr11_-_11925832 | 0.47 |
|
|
|
| chr21_-_2147085 | 0.47 |
ENSDART00000169262
|
zgc:163077
|
zgc:163077 |
| chr3_-_15584548 | 0.47 |
ENSDART00000137325
|
mvp
|
major vault protein |
| chr1_-_24988461 | 0.47 |
ENSDART00000054230
|
fgg
|
fibrinogen gamma chain |
| chr19_+_15538967 | 0.46 |
ENSDART00000171403
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr19_-_33624795 | 0.46 |
ENSDART00000109868
|
trib1
|
tribbles pseudokinase 1 |
| chr10_+_16267331 | 0.46 |
ENSDART00000129844
|
slc12a2
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
| chr20_+_29307142 | 0.46 |
ENSDART00000153016
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr3_+_22392439 | 0.45 |
|
|
|
| chr13_-_33574216 | 0.45 |
ENSDART00000065435
|
cst3
|
cystatin C (amyloid angiopathy and cerebral hemorrhage) |
| chr16_+_11138924 | 0.45 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr8_+_26273354 | 0.45 |
ENSDART00000053463
|
mgll
|
monoglyceride lipase |
| chr21_-_44570264 | 0.45 |
ENSDART00000159323
|
fundc2
|
fun14 domain containing 2 |
| chr16_+_24045707 | 0.45 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr23_-_4769238 | 0.44 |
ENSDART00000144536
|
cnbpa
|
CCHC-type zinc finger, nucleic acid binding protein a |
| chr10_-_210681 | 0.44 |
ENSDART00000059476
|
psmg1
|
proteasome (prosome, macropain) assembly chaperone 1 |
| chr14_-_943860 | 0.44 |
ENSDART00000010773
|
acsl1b
|
acyl-CoA synthetase long-chain family member 1b |
| chr3_+_49166063 | 0.44 |
ENSDART00000156347
|
epn3a
|
epsin 3a |
| chr16_+_22950567 | 0.44 |
ENSDART00000143957
|
flad1
|
flavin adenine dinucleotide synthetase 1 |
| chr11_-_5793 | 0.44 |
|
|
|
| chr25_-_10930008 | 0.43 |
ENSDART00000156846
|
mespbb
|
mesoderm posterior bb |
| KN150583v1_-_1389 | 0.43 |
|
|
|
| chr23_+_31036558 | 0.43 |
ENSDART00000115417
|
si:ch211-197l9.2
|
si:ch211-197l9.2 |
| chr16_+_49796978 | 0.43 |
ENSDART00000157100
|
ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr8_+_39630018 | 0.43 |
ENSDART00000125880
|
ENSDARG00000086589
|
ENSDARG00000086589 |
| chr11_+_11284217 | 0.43 |
ENSDART00000026814
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr4_-_71353374 | 0.43 |
|
|
|
| chr9_-_711269 | 0.42 |
ENSDART00000144625
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr16_+_11138879 | 0.42 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr10_-_42032702 | 0.42 |
|
|
|
| chr6_+_9185750 | 0.42 |
ENSDART00000161036
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr23_+_6043862 | 0.42 |
|
|
|
| chr8_-_31597252 | 0.42 |
ENSDART00000018886
|
ghra
|
growth hormone receptor a |
| chr7_-_49620185 | 0.42 |
ENSDART00000126240
|
cd44a
|
CD44 molecule (Indian blood group) a |
| chr3_-_32727588 | 0.42 |
ENSDART00000158916
|
si:dkey-16l2.20
|
si:dkey-16l2.20 |
| chr8_+_46319434 | 0.41 |
ENSDART00000145618
|
si:dkey-75a21.2
|
si:dkey-75a21.2 |
| chr14_+_6117282 | 0.41 |
ENSDART00000051556
|
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr21_+_20512024 | 0.41 |
ENSDART00000126005
|
efna5a
|
ephrin-A5a |
| chr22_+_9832516 | 0.41 |
ENSDART00000105942
|
si:dkey-253d23.3
|
si:dkey-253d23.3 |
| chr9_+_55999688 | 0.41 |
ENSDART00000172063
|
edar
|
ectodysplasin A receptor |
| chr10_-_20524860 | 0.40 |
|
|
|
| chr23_+_37754409 | 0.40 |
|
|
|
| chr5_+_17120453 | 0.39 |
|
|
|
| chr3_-_39245184 | 0.39 |
|
|
|
| chr11_-_23151247 | 0.39 |
|
|
|
| chr7_-_6273677 | 0.38 |
ENSDART00000173419
|
si:ch73-368j24.1
|
si:ch73-368j24.1 |
| chr12_-_28248133 | 0.38 |
ENSDART00000016283
|
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr3_+_21059221 | 0.38 |
ENSDART00000078807
|
zgc:123295
|
zgc:123295 |
| chr9_-_30560440 | 0.38 |
ENSDART00000079068
|
asb11
|
ankyrin repeat and SOCS box containing 11 |
| chr16_+_13928844 | 0.38 |
ENSDART00000090191
|
flcn
|
folliculin |
| KN149726v1_+_1094 | 0.38 |
|
|
|
| chr8_+_6533379 | 0.37 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr22_+_11745592 | 0.37 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr21_+_22808694 | 0.37 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr1_+_15827083 | 0.37 |
|
|
|
| chr4_-_15442828 | 0.37 |
ENSDART00000157414
|
plxna4
|
plexin A4 |
| chr21_+_8249235 | 0.37 |
ENSDART00000129749
|
psmb7
|
proteasome subunit beta 7 |
| chr7_+_26438049 | 0.36 |
ENSDART00000149426
|
cd82a
|
CD82 molecule a |
| chr1_+_50395721 | 0.36 |
ENSDART00000134065
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr8_-_337744 | 0.36 |
|
|
|
| chr14_-_5509554 | 0.36 |
|
|
|
| chr7_+_24762755 | 0.36 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr10_+_187740 | 0.36 |
ENSDART00000167367
|
ets2
|
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
| chr3_+_42380497 | 0.35 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr20_-_43826667 | 0.35 |
ENSDART00000100637
|
mixl1
|
Mix paired-like homeobox |
| chr5_-_34016383 | 0.35 |
ENSDART00000050271
ENSDART00000097975 |
hexb
|
hexosaminidase B (beta polypeptide) |
| chr25_-_22094023 | 0.35 |
ENSDART00000067478
|
pkp3a
|
plakophilin 3a |
| chr24_-_10928910 | 0.35 |
ENSDART00000127398
|
CR753886.1
|
ENSDARG00000090548 |
| chr19_-_7354071 | 0.35 |
ENSDART00000136528
|
rxrba
|
retinoid x receptor, beta a |
| chr11_-_13069266 | 0.35 |
ENSDART00000169052
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr4_+_5240888 | 0.35 |
ENSDART00000150391
|
si:ch211-214j24.14
|
si:ch211-214j24.14 |
| chr3_+_31793579 | 0.34 |
ENSDART00000127330
ENSDART00000126773 ENSDART00000055279 |
snrnp70
|
small nuclear ribonucleoprotein 70 (U1) |
| chr20_-_21773202 | 0.34 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr23_+_116397 | 0.34 |
|
|
|
| chr7_+_38408731 | 0.34 |
ENSDART00000007913
|
psmc3
|
proteasome 26S subunit, ATPase 3 |
| chr8_-_985673 | 0.34 |
ENSDART00000170737
|
smyd1b
|
SET and MYND domain containing 1b |
| chr5_-_66792947 | 0.34 |
ENSDART00000147009
|
si:dkey-251i10.2
|
si:dkey-251i10.2 |
| chr1_-_50147413 | 0.34 |
ENSDART00000080389
|
fam13a
|
family with sequence similarity 13, member A |
| chr24_-_9151388 | 0.33 |
ENSDART00000149875
|
tgif1
|
TGFB-induced factor homeobox 1 |
| chr7_+_34417030 | 0.33 |
ENSDART00000108473
|
plekhg4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr22_+_35113233 | 0.33 |
ENSDART00000123066
|
srfa
|
serum response factor a |
| chr17_+_134921 | 0.33 |
ENSDART00000166339
|
klhdc2
|
kelch domain containing 2 |
| chr4_-_72146177 | 0.33 |
ENSDART00000150546
|
si:dkey-262g12.3
|
si:dkey-262g12.3 |
| chr2_-_42011586 | 0.33 |
ENSDART00000045763
|
keap1a
|
kelch-like ECH-associated protein 1a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:1905178 | regulation of cardiac muscle tissue regeneration(GO:1905178) |
| 0.4 | 1.1 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.3 | 1.4 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.3 | 1.3 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.3 | 1.4 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.3 | 0.8 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.3 | 0.8 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.3 | 0.8 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.2 | 0.6 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.2 | 0.6 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.2 | 2.3 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 0.8 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.2 | 1.7 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.2 | 1.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.2 | 0.8 | GO:0003379 | establishment of cell polarity involved in gastrulation cell migration(GO:0003379) |
| 0.2 | 1.5 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.2 | 0.9 | GO:0032197 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.1 | 0.4 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.1 | 0.6 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.1 | 2.3 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.1 | 0.5 | GO:0019343 | homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) transsulfuration(GO:0019346) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.4 | GO:0046443 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.1 | 0.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.4 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.1 | GO:0048940 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.1 | 1.4 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.1 | 0.3 | GO:2000303 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.9 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.3 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 2.3 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 0.4 | GO:0010939 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) |
| 0.1 | 0.5 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 0.5 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.7 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.1 | 1.5 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 1.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.7 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 0.2 | GO:2000058 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.1 | 0.4 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.2 | GO:0035474 | selective angioblast sprouting(GO:0035474) |
| 0.1 | 0.5 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 0.2 | GO:0002280 | monocyte activation involved in immune response(GO:0002280) negative regulation of phagocytosis(GO:0050765) |
| 0.1 | 0.2 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.1 | 0.2 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.1 | 0.4 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.5 | GO:1902315 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.1 | 0.5 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.1 | 0.3 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.1 | 0.9 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.5 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.1 | 0.2 | GO:0036088 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.1 | 0.5 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 0.6 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.7 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.1 | 1.4 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 1.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.5 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 0.4 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.0 | 0.2 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.3 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.2 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.4 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.3 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.6 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.3 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.0 | 0.3 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.8 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.0 | 0.3 | GO:0090342 | regulation of cell aging(GO:0090342) |
| 0.0 | 0.2 | GO:0018158 | peptidyl-lysine oxidation(GO:0018057) protein oxidation(GO:0018158) |
| 0.0 | 0.0 | GO:2000391 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.0 | 0.3 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.1 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.5 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.2 | GO:1900024 | regulation of substrate adhesion-dependent cell spreading(GO:1900024) |
| 0.0 | 0.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.1 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.2 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 0.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 1.2 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.4 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.2 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.6 | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030947) |
| 0.0 | 0.2 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.3 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.6 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.9 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.5 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.4 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.8 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.0 | 0.1 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.0 | 0.1 | GO:0000050 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.0 | 0.3 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 2.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0072149 | visceral serous pericardium development(GO:0061032) glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.0 | 0.1 | GO:0071939 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.8 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.3 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0046099 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine metabolic process(GO:0046098) guanine biosynthetic process(GO:0046099) hypoxanthine metabolic process(GO:0046100) |
| 0.0 | 0.1 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.3 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.0 | 0.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.1 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.4 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.1 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.1 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.4 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 0.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.5 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.1 | GO:0050482 | icosanoid secretion(GO:0032309) acid secretion(GO:0046717) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.2 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.2 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.5 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 2.5 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 0.7 | GO:0034620 | cellular response to unfolded protein(GO:0034620) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.9 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.7 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.4 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.0 | 0.9 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.3 | 0.8 | GO:0098594 | mucin granule(GO:0098594) |
| 0.3 | 1.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 2.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 0.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 0.7 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 0.4 | GO:0031362 | intrinsic component of external side of plasma membrane(GO:0031233) anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.4 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 2.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.3 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 1.9 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.9 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 1.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 2.6 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.3 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 1.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 1.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.3 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.3 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 1.9 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.2 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 1.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 2.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 1.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.4 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 1.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.4 | 1.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.3 | 1.0 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.3 | 0.8 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.2 | 1.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.2 | 0.9 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.2 | 0.8 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.2 | 3.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 0.6 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.2 | 1.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 0.4 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.1 | 1.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.5 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 0.4 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.1 | 1.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 2.1 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.9 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.3 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.1 | 0.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.9 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 0.4 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.6 | GO:0004675 | transmembrane receptor protein serine/threonine kinase activity(GO:0004675) |
| 0.1 | 0.4 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.5 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.6 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.2 | GO:0043121 | neurotrophin binding(GO:0043121) |
| 0.1 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.5 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 2.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 1.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0034417 | bisphosphoglycerate 3-phosphatase activity(GO:0034417) |
| 0.0 | 0.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 2.5 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.3 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.4 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) retinol transporter activity(GO:0034632) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.0 | 0.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.0 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.0 | GO:0004044 | amidophosphoribosyltransferase activity(GO:0004044) |
| 0.0 | 0.7 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 1.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.3 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 1.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 2.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 0.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 1.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.8 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.2 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.8 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 2.3 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 2.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 0.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.9 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |