Project
DANIO-CODE
Navigation
Downloads
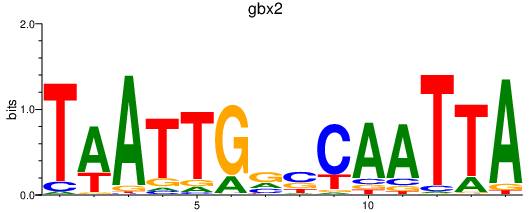
Results for gbx2
Z-value: 1.32
Transcription factors associated with gbx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gbx2
|
ENSDARG00000002933 | gastrulation brain homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gbx2 | dr10_dc_chr6_+_15904220_15904239 | -0.76 | 5.7e-04 | Click! |
Activity profile of gbx2 motif
Sorted Z-values of gbx2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of gbx2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_6442588 | 7.05 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr21_-_32747592 | 5.45 |
|
|
|
| chr1_-_18118467 | 4.12 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr19_-_10411573 | 3.63 |
ENSDART00000171232
|
ccdc106b
|
coiled-coil domain containing 106b |
| chr24_-_26915897 | 3.34 |
ENSDART00000142864
|
stag1b
|
stromal antigen 1b |
| chr7_+_45747622 | 3.26 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr14_+_50010976 | 3.22 |
ENSDART00000171955
|
CR855307.1
|
ENSDARG00000099628 |
| chr5_-_19619115 | 2.88 |
ENSDART00000026516
|
pxmp2
|
peroxisomal membrane protein 2 |
| chr7_+_45747395 | 2.84 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr14_-_49777408 | 2.77 |
ENSDART00000166463
|
cnot6b
|
CCR4-NOT transcription complex, subunit 6b |
| chr15_-_16241412 | 2.56 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr5_-_19619201 | 2.48 |
ENSDART00000026516
|
pxmp2
|
peroxisomal membrane protein 2 |
| chr1_+_18118735 | 2.34 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| KN149710v1_+_38638 | 2.29 |
|
|
|
| chr8_+_23083842 | 2.22 |
ENSDART00000025171
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr25_-_26399777 | 2.09 |
ENSDART00000140643
|
commd4
|
COMM domain containing 4 |
| chr14_+_23420053 | 2.09 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr6_+_3556296 | 2.08 |
ENSDART00000041627
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr11_-_12174576 | 1.99 |
ENSDART00000147670
|
npepps
|
aminopeptidase puromycin sensitive |
| chr17_+_24300451 | 1.99 |
ENSDART00000064083
|
otx1b
|
orthodenticle homeobox 1b |
| chr11_-_6442547 | 1.97 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr7_+_59371957 | 1.86 |
ENSDART00000125570
|
trmt44
|
tRNA methyltransferase 44 homolog (S. cerevisiae) |
| chr6_-_42371527 | 1.85 |
ENSDART00000039868
|
usp4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr15_+_35076414 | 1.82 |
ENSDART00000165210
|
zgc:66024
|
zgc:66024 |
| chr11_-_6442490 | 1.82 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr19_-_10411518 | 1.75 |
ENSDART00000171232
|
ccdc106b
|
coiled-coil domain containing 106b |
| chr24_-_32668102 | 1.71 |
ENSDART00000066590
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr20_+_54404987 | 1.71 |
ENSDART00000099338
|
actr10
|
ARP10 actin related protein 10 homolog |
| chr20_-_7079663 | 1.70 |
ENSDART00000040793
|
sirt5
|
sirtuin 5 |
| chr19_-_25565317 | 1.65 |
ENSDART00000175266
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr8_+_23084130 | 1.65 |
ENSDART00000025171
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr21_-_32747544 | 1.62 |
|
|
|
| chr20_-_7079412 | 1.54 |
ENSDART00000040793
|
sirt5
|
sirtuin 5 |
| chr22_-_9832436 | 1.50 |
|
|
|
| chr15_-_16241500 | 1.47 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr24_-_26915972 | 1.45 |
ENSDART00000142864
|
stag1b
|
stromal antigen 1b |
| chr22_+_17803347 | 1.44 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr20_-_7079949 | 1.37 |
ENSDART00000040793
|
sirt5
|
sirtuin 5 |
| chr10_-_14971565 | 1.32 |
ENSDART00000147653
|
smad2
|
SMAD family member 2 |
| chr19_-_10411878 | 1.23 |
ENSDART00000136653
|
ccdc106b
|
coiled-coil domain containing 106b |
| chr8_-_17974364 | 1.19 |
ENSDART00000133666
|
acot11b
|
acyl-CoA thioesterase 11b |
| chr24_-_33915389 | 1.15 |
ENSDART00000079202
|
abcb8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr8_-_17974316 | 1.15 |
ENSDART00000122730
|
acot11b
|
acyl-CoA thioesterase 11b |
| chr22_-_9832520 | 1.14 |
|
|
|
| chr2_+_42077559 | 1.14 |
ENSDART00000023208
|
ENSDARG00000014209
|
ENSDARG00000014209 |
| chr5_-_62982537 | 1.08 |
ENSDART00000111403
|
ciz1b
|
cdkn1a interacting zinc finger protein 1b |
| chr8_-_50151276 | 1.05 |
|
|
|
| chr15_+_34584015 | 1.03 |
|
|
|
| chr16_+_30393572 | 1.02 |
|
|
|
| chr5_-_62982637 | 1.02 |
ENSDART00000111403
|
ciz1b
|
cdkn1a interacting zinc finger protein 1b |
| chr17_+_24300332 | 0.98 |
ENSDART00000064083
|
otx1b
|
orthodenticle homeobox 1b |
| chr20_-_7079708 | 0.97 |
ENSDART00000040793
|
sirt5
|
sirtuin 5 |
| chr19_-_25564720 | 0.92 |
ENSDART00000148432
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr14_+_50011161 | 0.89 |
ENSDART00000171955
|
CR855307.1
|
ENSDARG00000099628 |
| chr23_+_4315077 | 0.83 |
ENSDART00000008761
|
arl6ip5b
|
ADP-ribosylation factor-like 6 interacting protein 5b |
| chr18_+_31078854 | 0.74 |
ENSDART00000164585
ENSDART00000159316 |
mvda
|
mevalonate (diphospho) decarboxylase a |
| chr11_-_29520809 | 0.74 |
ENSDART00000079117
|
plekha3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr5_+_38225036 | 0.73 |
ENSDART00000076835
|
mrpl1
|
mitochondrial ribosomal protein L1 |
| chr10_-_14971401 | 0.69 |
ENSDART00000044756
ENSDART00000128579 |
smad2
|
SMAD family member 2 |
| chr20_+_54404819 | 0.62 |
ENSDART00000099338
|
actr10
|
ARP10 actin related protein 10 homolog |
| chr21_-_32747483 | 0.58 |
|
|
|
| chr2_-_53797678 | 0.54 |
ENSDART00000057053
|
im:7138239
|
im:7138239 |
| chr16_-_26982451 | 0.52 |
ENSDART00000078119
|
ino80c
|
INO80 complex subunit C |
| chr2_-_53797472 | 0.51 |
ENSDART00000057053
|
im:7138239
|
im:7138239 |
| chr25_+_14153652 | 0.50 |
|
|
|
| chr16_-_22378805 | 0.47 |
ENSDART00000124718
|
aqp10a
|
aquaporin 10a |
| chr8_+_23083870 | 0.46 |
ENSDART00000025171
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr6_-_1638631 | 0.45 |
ENSDART00000087039
|
ENSDARG00000015563
|
ENSDARG00000015563 |
| chr7_-_45747162 | 0.42 |
ENSDART00000172591
|
zgc:162297
|
zgc:162297 |
| chr24_+_33916364 | 0.41 |
ENSDART00000136040
|
atg9b
|
autophagy related 9B |
| chr3_+_22386709 | 0.38 |
|
|
|
| chr11_+_16082265 | 0.37 |
ENSDART00000081035
|
slc25a26
|
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr16_-_36085222 | 0.37 |
ENSDART00000159134
|
OSCP1
|
organic solute carrier partner 1 |
| chr18_+_15964146 | 0.36 |
|
|
|
| chr13_-_199651 | 0.36 |
ENSDART00000165165
|
ddx21
|
DEAD (Asp-Glu-Ala-Asp) box helicase 21 |
| chr6_-_42371446 | 0.34 |
ENSDART00000039868
|
usp4
|
ubiquitin specific peptidase 4 (proto-oncogene) |
| chr2_-_53797503 | 0.34 |
ENSDART00000057053
|
im:7138239
|
im:7138239 |
| chr13_-_33545295 | 0.34 |
ENSDART00000020350
|
snx5
|
sorting nexin 5 |
| chr5_-_25133456 | 0.33 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr15_+_44514665 | 0.32 |
|
|
|
| chr7_-_45747112 | 0.30 |
ENSDART00000172591
|
zgc:162297
|
zgc:162297 |
| chr6_-_1638559 | 0.22 |
ENSDART00000087039
|
ENSDARG00000015563
|
ENSDARG00000015563 |
| chr18_+_7680416 | 0.16 |
ENSDART00000132369
|
rabl2
|
RAB, member of RAS oncogene family-like 2 |
| chr2_+_35612974 | 0.15 |
ENSDART00000098734
|
cacybp
|
calcyclin binding protein |
| chr7_-_33413478 | 0.11 |
ENSDART00000130553
|
tle3b
|
transducin-like enhancer of split 3b |
| chr17_+_41313762 | 0.10 |
ENSDART00000128482
|
fosl2
|
fos-like antigen 2 |
| chr14_-_33010712 | 0.05 |
ENSDART00000010019
|
upf3b
|
UPF3 regulator of nonsense transcripts homolog B (yeast) |
| chr22_+_2074020 | 0.04 |
ENSDART00000106540
|
znf1161
|
zinc finger protein 1161 |
| chr25_-_20426215 | 0.04 |
|
|
|
| chr11_-_12174627 | 0.03 |
ENSDART00000147670
|
npepps
|
aminopeptidase puromycin sensitive |
| chr19_-_42759922 | 0.01 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.6 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) protein demalonylation(GO:0036046) peptidyl-lysine demalonylation(GO:0036047) protein desuccinylation(GO:0036048) peptidyl-lysine desuccinylation(GO:0036049) protein deglutarylation(GO:0061698) |
| 0.5 | 2.3 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.3 | 4.3 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.3 | 2.0 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 0.7 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.2 | 1.5 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.1 | 0.7 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 2.0 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 6.1 | GO:0000082 | G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.1 | 4.8 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.1 | 2.8 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 0.4 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 1.9 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 2.1 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.1 | GO:1905207 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.0 | 2.3 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.7 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.8 | GO:0051051 | negative regulation of transport(GO:0051051) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.1 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.4 | 4.8 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.2 | 2.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 2.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 2.0 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.5 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 2.1 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 11.1 | GO:0005739 | mitochondrion(GO:0005739) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.6 | GO:0036055 | protein-malonyllysine demalonylase activity(GO:0036054) protein-succinyllysine desuccinylase activity(GO:0036055) protein-glutaryllysine deglutarylase activity(GO:0061697) |
| 0.6 | 1.9 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.6 | 2.3 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.5 | 4.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.3 | 2.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 2.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.2 | 0.7 | GO:1902388 | ceramide transporter activity(GO:0035620) sphingolipid transporter activity(GO:0046624) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.2 | 0.5 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 2.0 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 2.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 6.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.4 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.5 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 4.8 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 1.2 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 2.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 5.6 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 1.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.1 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 2.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 2.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |