Project
DANIO-CODE
Navigation
Downloads
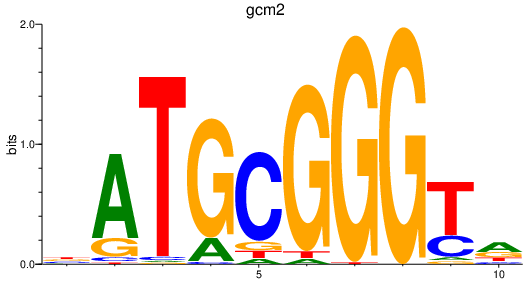
Results for gcm2
Z-value: 1.06
Transcription factors associated with gcm2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gcm2
|
ENSDARG00000045413 | glial cells missing transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gcm2 | dr10_dc_chr24_-_8691595_8691597 | 0.53 | 3.4e-02 | Click! |
Activity profile of gcm2 motif
Sorted Z-values of gcm2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of gcm2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_39187544 | 3.08 |
ENSDART00000148648
|
rps25
|
ribosomal protein S25 |
| chr7_-_6219627 | 2.51 |
ENSDART00000172898
|
HIST2H2AB (1 of many)
|
zgc:165551 |
| chr25_+_35791595 | 1.86 |
ENSDART00000103013
|
ENSDARG00000070286
|
ENSDARG00000070286 |
| chr20_+_17840364 | 1.71 |
ENSDART00000024627
|
cdh2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr16_-_20506304 | 1.57 |
ENSDART00000134980
|
si:dkeyp-86h10.3
|
si:dkeyp-86h10.3 |
| chr16_+_25491981 | 1.56 |
ENSDART00000086333
|
jarid2a
|
jumonji, AT rich interactive domain 2a |
| chr5_-_15774816 | 1.46 |
ENSDART00000090684
|
kremen1
|
kringle containing transmembrane protein 1 |
| chr1_-_20218263 | 1.41 |
ENSDART00000078271
|
cpe
|
carboxypeptidase E |
| chr25_+_35798656 | 1.40 |
ENSDART00000103006
|
zgc:110434
|
zgc:110434 |
| chr25_-_35817806 | 1.37 |
ENSDART00000129969
|
ENSDARG00000086604
|
ENSDARG00000086604 |
| chr7_-_6295302 | 1.33 |
ENSDART00000173199
|
HIST2H2AB (1 of many)
|
si:ch1073-153i20.5 |
| chr25_+_35778740 | 1.31 |
ENSDART00000073404
|
zgc:114037
|
zgc:114037 |
| chr25_+_34628342 | 1.30 |
ENSDART00000155501
|
zgc:110434
|
zgc:110434 |
| chr17_-_35934175 | 1.28 |
ENSDART00000110040
ENSDART00000137525 |
sox11a
AL929378.1
|
SRY (sex determining region Y)-box 11a ENSDARG00000092907 |
| chr1_-_50513645 | 1.23 |
ENSDART00000047851
|
jag1a
|
jagged 1a |
| chr6_+_42821679 | 1.21 |
ENSDART00000046498
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr7_-_26035308 | 1.21 |
ENSDART00000131906
|
zgc:77439
|
zgc:77439 |
| chr25_-_34635772 | 1.17 |
ENSDART00000113870
|
ENSDARG00000075379
|
ENSDARG00000075379 |
| chr16_+_32605735 | 1.16 |
ENSDART00000093250
|
pou3f2b
|
POU class 3 homeobox 2b |
| chr25_+_35787139 | 1.16 |
ENSDART00000073402
|
si:ch211-113a14.16
|
si:ch211-113a14.16 |
| chr1_-_50513873 | 1.09 |
ENSDART00000137172
|
jag1a
|
jagged 1a |
| chr7_-_6295176 | 1.06 |
ENSDART00000173199
|
HIST2H2AB (1 of many)
|
si:ch1073-153i20.5 |
| chr17_-_26592356 | 1.00 |
ENSDART00000016608
|
mrpl57
|
mitochondrial ribosomal protein L57 |
| chr1_+_4704661 | 0.99 |
ENSDART00000040204
|
tuba8l2
|
tubulin, alpha 8 like 2 |
| chr16_+_53178713 | 0.98 |
ENSDART00000174634
|
CABZ01062546.1
|
ENSDARG00000108858 |
| chr7_+_1397973 | 0.97 |
|
|
|
| chr25_-_33920256 | 0.95 |
|
|
|
| chr16_+_12376719 | 0.95 |
ENSDART00000060037
|
ENSDARG00000040971
|
ENSDARG00000040971 |
| chr4_+_317946 | 0.90 |
ENSDART00000132625
|
tulp4a
|
tubby like protein 4a |
| chr9_+_26292183 | 0.88 |
|
|
|
| chr18_+_41243277 | 0.85 |
|
|
|
| chr14_+_6834563 | 0.84 |
ENSDART00000170994
ENSDART00000129898 |
ctsf
|
cathepsin F |
| chr11_+_23522743 | 0.83 |
ENSDART00000121874
|
nfasca
|
neurofascin homolog (chicken) a |
| chr2_-_32521879 | 0.79 |
ENSDART00000056639
|
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr20_+_27048717 | 0.78 |
ENSDART00000062066
|
si:dkey-177p2.6
|
si:dkey-177p2.6 |
| chr23_+_1675422 | 0.78 |
|
|
|
| chr9_-_56292487 | 0.78 |
ENSDART00000151720
|
si:ch211-39i22.1
|
si:ch211-39i22.1 |
| chr3_-_58488929 | 0.74 |
ENSDART00000042386
|
unm_sa1261
|
un-named sa1261 |
| chr19_+_2013850 | 0.73 |
ENSDART00000048850
|
sh3bp5a
|
SH3-domain binding protein 5a (BTK-associated) |
| chr6_-_37644217 | 0.72 |
ENSDART00000065119
|
chst7
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
| chr7_+_72532825 | 0.72 |
ENSDART00000175316
|
FO818686.1
|
ENSDARG00000108531 |
| chr18_-_21229799 | 0.71 |
ENSDART00000060160
|
calb2a
|
calbindin 2a |
| chr5_-_41645167 | 0.70 |
|
|
|
| chr5_+_18421698 | 0.70 |
ENSDART00000051621
|
pgam5
|
PGAM family member 5, serine/threonine protein phosphatase, mitochondrial |
| chr21_+_27241416 | 0.68 |
|
|
|
| chr9_-_3429153 | 0.68 |
ENSDART00000111386
|
dlx2a
|
distal-less homeobox 2a |
| chr17_-_5453357 | 0.67 |
ENSDART00000004043
|
enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr16_+_5778398 | 0.67 |
ENSDART00000011166
ENSDART00000126445 |
zgc:158689
|
zgc:158689 |
| chr1_-_44239584 | 0.66 |
ENSDART00000137216
|
tmem176
|
transmembrane protein 176 |
| chr13_-_24779651 | 0.63 |
ENSDART00000077775
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr1_+_1964902 | 0.63 |
ENSDART00000138396
|
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr16_+_55008251 | 0.60 |
ENSDART00000176995
|
CABZ01087388.1
|
ENSDARG00000106538 |
| chr14_-_15393962 | 0.60 |
ENSDART00000161123
ENSDART00000167146 |
neurl1b
|
neuralized E3 ubiquitin protein ligase 1B |
| chr22_-_4622401 | 0.60 |
ENSDART00000106166
|
rx1
|
retinal homeobox gene 1 |
| chr25_-_21796677 | 0.59 |
ENSDART00000089642
|
fbxo31
|
F-box protein 31 |
| chr25_-_34639606 | 0.58 |
ENSDART00000126259
|
zgc:110434
|
zgc:110434 |
| chr12_-_7790348 | 0.57 |
ENSDART00000148673
|
ank3b
|
ankyrin 3b |
| chr1_-_53746151 | 0.57 |
ENSDART00000179565
|
CABZ01043826.1
|
ENSDARG00000108130 |
| chr1_+_33563354 | 0.57 |
ENSDART00000075584
|
kctd12.2
|
potassium channel tetramerisation domain containing 12.2 |
| chr23_+_12799718 | 0.55 |
|
|
|
| chr14_+_7393010 | 0.55 |
ENSDART00000123139
|
brd8
|
bromodomain containing 8 |
| chr4_-_71770315 | 0.55 |
ENSDART00000171434
|
zgc:162958
|
zgc:162958 |
| chr24_-_6129575 | 0.54 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr6_+_59749159 | 0.54 |
|
|
|
| chr17_+_51272621 | 0.52 |
|
|
|
| chr3_+_39617522 | 0.52 |
|
|
|
| chr7_+_73600230 | 0.52 |
ENSDART00000113375
|
HIST2H2AB (1 of many)
|
histone cluster 2 H2A family member b |
| chr24_-_26339981 | 0.51 |
ENSDART00000135496
|
eif5a
|
eukaryotic translation initiation factor 5A |
| chr7_-_73632917 | 0.51 |
ENSDART00000122392
|
HIST2H2AB (1 of many)
|
histone cluster 2 H2A family member b |
| chr14_-_30747259 | 0.51 |
ENSDART00000002250
|
hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr5_+_69490090 | 0.49 |
|
|
|
| chr22_-_37676744 | 0.49 |
ENSDART00000028085
|
ttc14
|
tetratricopeptide repeat domain 14 |
| chr10_+_29965951 | 0.49 |
ENSDART00000116893
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr5_+_37996820 | 0.47 |
ENSDART00000015136
|
mogat3b
|
monoacylglycerol O-acyltransferase 3b |
| chr15_+_8359658 | 0.46 |
ENSDART00000152603
|
egln2
|
egl-9 family hypoxia-inducible factor 2 |
| chr22_-_20316908 | 0.46 |
ENSDART00000165667
|
tcf3b
|
transcription factor 3b |
| chr13_-_42986821 | 0.45 |
ENSDART00000164439
|
si:ch211-106f21.1
|
si:ch211-106f21.1 |
| chr24_+_24924379 | 0.44 |
ENSDART00000115165
|
amer2
|
APC membrane recruitment protein 2 |
| chr18_-_17410180 | 0.43 |
ENSDART00000060949
|
zfpm1
|
zinc finger protein, FOG family member 1 |
| chr18_+_2991098 | 0.42 |
ENSDART00000165048
|
aamdc
|
adipogenesis associated, Mth938 domain containing |
| chr14_+_44399056 | 0.42 |
ENSDART00000173043
|
atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
| chr7_-_20201358 | 0.41 |
ENSDART00000174001
|
ntn5
|
netrin 5 |
| chr5_-_25509259 | 0.41 |
ENSDART00000137376
|
BX510923.1
|
ENSDARG00000094488 |
| chr13_-_36420228 | 0.39 |
|
|
|
| chr3_-_39208943 | 0.39 |
|
|
|
| chr17_-_5453436 | 0.38 |
ENSDART00000004043
|
enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr19_+_32679320 | 0.38 |
|
|
|
| chr19_+_32679258 | 0.38 |
|
|
|
| chr17_+_13088982 | 0.38 |
ENSDART00000012670
|
pnn
|
pinin, desmosome associated protein |
| chr1_-_50513762 | 0.37 |
ENSDART00000047851
|
jag1a
|
jagged 1a |
| chr10_-_20399472 | 0.33 |
ENSDART00000080143
|
sfrp1b
|
secreted frizzled-related protein 1b |
| chr5_-_25509188 | 0.33 |
ENSDART00000137376
|
BX510923.1
|
ENSDARG00000094488 |
| chr25_-_34635697 | 0.33 |
ENSDART00000113870
|
ENSDARG00000075379
|
ENSDARG00000075379 |
| chr4_+_21319791 | 0.33 |
|
|
|
| chr16_-_52761159 | 0.32 |
ENSDART00000111869
|
ubr5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr25_-_35817715 | 0.32 |
ENSDART00000129969
|
ENSDARG00000086604
|
ENSDARG00000086604 |
| chr19_+_4093543 | 0.30 |
ENSDART00000172204
|
meaf6
|
MYST/Esa1-associated factor 6 |
| chr3_+_32360862 | 0.30 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr5_-_25509310 | 0.29 |
ENSDART00000137376
|
BX510923.1
|
ENSDARG00000094488 |
| chr7_-_26035541 | 0.29 |
ENSDART00000057288
|
zgc:77439
|
zgc:77439 |
| chr15_+_29190162 | 0.28 |
ENSDART00000105222
|
zgc:101731
|
zgc:101731 |
| chr13_+_14873339 | 0.27 |
ENSDART00000057810
|
emx1
|
empty spiracles homeobox 1 |
| chr14_-_16791447 | 0.27 |
ENSDART00000158002
|
CR812832.1
|
ENSDARG00000103278 |
| chr23_+_32085181 | 0.27 |
ENSDART00000053509
|
plagx
|
pleiomorphic adenoma gene X |
| chr22_-_8228763 | 0.26 |
ENSDART00000097196
|
zmp:0000001043
|
zmp:0000001043 |
| chr16_-_22260272 | 0.26 |
ENSDART00000138419
|
CR855277.1
|
ENSDARG00000092020 |
| chr9_-_29850713 | 0.25 |
ENSDART00000156609
|
CU855930.2
|
ENSDARG00000097441 |
| chr1_+_44239727 | 0.25 |
ENSDART00000141145
|
si:dkey-9i23.16
|
si:dkey-9i23.16 |
| chr1_+_3674819 | 0.25 |
|
|
|
| chr4_+_19545842 | 0.24 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr20_+_23392782 | 0.24 |
|
|
|
| chr11_+_35790782 | 0.22 |
ENSDART00000125221
|
CR933559.1
|
ENSDARG00000087939 |
| chr2_-_24614261 | 0.21 |
ENSDART00000099532
|
vmhc
|
ventricular myosin heavy chain |
| chr6_-_6091628 | 0.21 |
ENSDART00000132350
|
rtn4a
|
reticulon 4a |
| chr20_+_43751193 | 0.21 |
ENSDART00000017269
|
parp1
|
poly (ADP-ribose) polymerase 1 |
| chr6_+_15635696 | 0.21 |
ENSDART00000128939
|
iqca1
|
IQ motif containing with AAA domain 1 |
| chr22_+_26840335 | 0.20 |
ENSDART00000158756
|
crebbpa
|
CREB binding protein a |
| chr7_-_20201693 | 0.20 |
ENSDART00000174001
|
ntn5
|
netrin 5 |
| chr17_+_23534824 | 0.19 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr14_-_2295383 | 0.18 |
ENSDART00000162537
ENSDART00000106705 |
pcdh2ab3
pcdh2ab3
|
protocadherin 2 alpha b 3 protocadherin 2 alpha b 3 |
| chr3_+_32618086 | 0.17 |
ENSDART00000125260
|
hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase |
| chr22_-_20317201 | 0.17 |
ENSDART00000161610
|
tcf3b
|
transcription factor 3b |
| chr20_-_37733350 | 0.17 |
|
|
|
| chr25_-_3730472 | 0.17 |
|
|
|
| chr16_-_32718222 | 0.17 |
ENSDART00000161395
|
pnisr
|
PNN-interacting serine/arginine-rich protein |
| chr18_+_8388897 | 0.17 |
ENSDART00000083421
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr5_+_69489874 | 0.16 |
|
|
|
| chr17_-_25545464 | 0.16 |
ENSDART00000040032
ENSDART00000163447 |
rtn4ip1
|
reticulon 4 interacting protein 1 |
| chr7_-_4987762 | 0.15 |
ENSDART00000097877
|
ltb4r2a
|
leukotriene B4 receptor 2a |
| KN150118v1_+_46683 | 0.15 |
|
|
|
| chr22_-_20317131 | 0.15 |
ENSDART00000161610
|
tcf3b
|
transcription factor 3b |
| chr24_+_16005004 | 0.14 |
ENSDART00000163086
ENSDART00000152087 |
si:dkey-118j18.2
|
si:dkey-118j18.2 |
| chr6_+_42821484 | 0.13 |
ENSDART00000046498
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr3_-_31111443 | 0.13 |
ENSDART00000076925
|
ITPRIPL2
|
ITPRIP like 2 |
| chr11_+_38013564 | 0.13 |
ENSDART00000171496
|
CDK18
|
cyclin dependent kinase 18 |
| chr16_+_12376861 | 0.12 |
ENSDART00000060037
|
ENSDARG00000040971
|
ENSDARG00000040971 |
| chr13_+_30546398 | 0.12 |
|
|
|
| chr7_-_6219521 | 0.12 |
ENSDART00000161304
|
HIST2H2AB (1 of many)
|
zgc:165551 |
| chr7_+_5786385 | 0.12 |
ENSDART00000173404
|
HIST2H2AB (1 of many)
|
histone cluster 2 H2A family member b |
| chr6_+_46404650 | 0.11 |
ENSDART00000131203
ENSDART00000103472 ENSDART00000132845 ENSDART00000138567 ENSDART00000168440 |
pbrm1l
|
polybromo 1, like |
| chr23_-_31033917 | 0.10 |
ENSDART00000142661
|
osbpl2a
|
oxysterol binding protein-like 2a |
| chr6_-_60172823 | 0.09 |
|
|
|
| chr11_+_26138359 | 0.09 |
ENSDART00000087652
|
cpne1
|
copine I |
| chr10_+_15078155 | 0.08 |
|
|
|
| chr6_+_19476202 | 0.07 |
ENSDART00000113911
|
micall1a
|
MICAL-like 1a |
| chr6_+_15635738 | 0.07 |
ENSDART00000128939
|
iqca1
|
IQ motif containing with AAA domain 1 |
| chr11_-_41798903 | 0.05 |
ENSDART00000162944
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr7_-_73632999 | 0.05 |
ENSDART00000122392
|
HIST2H2AB (1 of many)
|
histone cluster 2 H2A family member b |
| chr20_-_54645903 | 0.04 |
ENSDART00000038745
|
yy1b
|
YY1 transcription factor b |
| chr5_-_15774729 | 0.04 |
ENSDART00000090684
|
kremen1
|
kringle containing transmembrane protein 1 |
| chr21_-_19649065 | 0.04 |
ENSDART00000131562
ENSDART00000133467 |
BX927394.1
|
ENSDARG00000092538 |
| chr16_+_51598269 | 0.03 |
ENSDART00000169443
|
SLC9A1
|
solute carrier family 9 member A1 |
| chr13_-_25153833 | 0.02 |
ENSDART00000039828
ENSDART00000110304 |
vcla
|
vinculin a |
| chr22_-_29810429 | 0.02 |
ENSDART00000135503
|
BX649294.1
|
ENSDARG00000092881 |
| chr7_-_18894589 | 0.01 |
ENSDART00000142924
|
kirrel1a
|
kin of IRRE like a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0098743 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.3 | 1.5 | GO:0048901 | anterior lateral line neuromast development(GO:0048901) anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.3 | 1.1 | GO:0050820 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.2 | 0.7 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.2 | 0.7 | GO:1902869 | negative regulation of photoreceptor cell differentiation(GO:0046533) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.2 | 0.5 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.4 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 0.5 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.1 | 1.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 2.7 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.1 | 0.6 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 0.3 | GO:0002706 | regulation of lymphocyte mediated immunity(GO:0002706) |
| 0.1 | 0.4 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.1 | 0.7 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.8 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.1 | 0.8 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.3 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.7 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.3 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.1 | 0.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.7 | GO:0061099 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.3 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.5 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.6 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 1.2 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.2 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.0 | 0.1 | GO:0001961 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.0 | 1.3 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.6 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.3 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.4 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 1.4 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.5 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.8 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.7 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.6 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.2 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 1.4 | GO:0016485 | protein processing(GO:0016485) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 3.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 1.0 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.5 | GO:0005643 | nuclear pore(GO:0005643) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.3 | 1.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 1.6 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.2 | 0.7 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 2.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.5 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 1.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.5 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.2 | GO:0004953 | icosanoid receptor activity(GO:0004953) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.5 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 1.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.7 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.2 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 4.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 1.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 3.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 0.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |