Project
DANIO-CODE
Navigation
Downloads
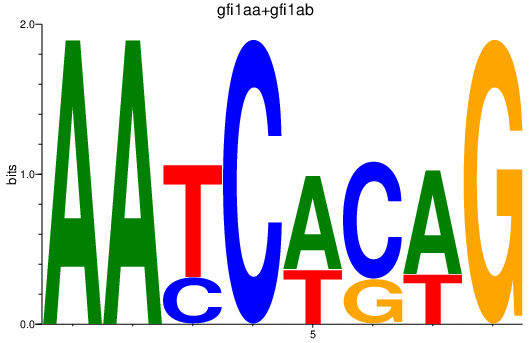
Results for gfi1aa+gfi1ab
Z-value: 1.08
Transcription factors associated with gfi1aa+gfi1ab
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gfi1aa
|
ENSDARG00000020746 | growth factor independent 1A transcription repressor a |
|
gfi1ab
|
ENSDARG00000044457 | growth factor independent 1A transcription repressor b |
Activity profile of gfi1aa+gfi1ab motif
Sorted Z-values of gfi1aa+gfi1ab motif
Network of associatons between targets according to the STRING database.
First level regulatory network of gfi1aa+gfi1ab
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| KN149735v1_-_1679 | 2.35 |
|
|
|
| chr12_-_28680035 | 1.01 |
ENSDART00000020667
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr3_-_32686790 | 1.01 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr1_+_30840656 | 0.97 |
ENSDART00000075286
|
slc2a15b
|
solute carrier family 2 (facilitated glucose transporter), member 15b |
| chr13_+_15685540 | 0.96 |
ENSDART00000137061
|
klc1a
|
kinesin light chain 1a |
| chr1_+_30840735 | 0.96 |
ENSDART00000075286
|
slc2a15b
|
solute carrier family 2 (facilitated glucose transporter), member 15b |
| chr16_+_17857126 | 0.95 |
ENSDART00000149408
|
them4
|
thioesterase superfamily member 4 |
| chr17_+_15425559 | 0.95 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr13_-_8360779 | 0.94 |
ENSDART00000058107
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr6_+_49772891 | 0.94 |
ENSDART00000134207
|
ctsz
|
cathepsin Z |
| chr10_-_33435736 | 0.94 |
ENSDART00000023509
|
ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr18_-_20476969 | 0.90 |
ENSDART00000060311
|
paqr5a
|
progestin and adipoQ receptor family member Va |
| chr10_+_38720500 | 0.88 |
ENSDART00000067448
|
acat1
|
acetyl-CoA acetyltransferase 1 |
| chr20_+_31524061 | 0.85 |
|
|
|
| chr24_+_17190237 | 0.83 |
ENSDART00000126435
|
pdia4
|
protein disulfide isomerase family A, member 4 |
| chr17_-_21046246 | 0.81 |
ENSDART00000078763
|
vsx1
|
visual system homeobox 1 homolog, chx10-like |
| chr5_-_31173320 | 0.80 |
ENSDART00000122066
|
fam102ab
|
family with sequence similarity 102, member Ab |
| chr4_-_20456825 | 0.79 |
ENSDART00000003621
ENSDART00000132356 |
sinup
|
siaz-interacting nuclear protein |
| chr21_+_26660833 | 0.78 |
ENSDART00000004109
|
gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr15_-_24934442 | 0.76 |
ENSDART00000127047
|
tusc5a
|
tumor suppressor candidate 5a |
| chr4_+_9278836 | 0.73 |
ENSDART00000014897
|
srgap1b
|
SLIT-ROBO Rho GTPase activating protein 1b |
| chr13_-_36719000 | 0.72 |
ENSDART00000129562
ENSDART00000150899 |
nin
|
ninein (GSK3B interacting protein) |
| chr1_-_45958767 | 0.71 |
ENSDART00000053222
|
rcbtb1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
| chr23_-_31719203 | 0.70 |
ENSDART00000148122
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr10_+_42111560 | 0.70 |
ENSDART00000126248
|
tmem120b
|
transmembrane protein 120B |
| chr13_-_42410874 | 0.70 |
ENSDART00000146843
|
btaf1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated |
| chr11_-_40383013 | 0.69 |
ENSDART00000112140
|
fam213b
|
family with sequence similarity 213, member B |
| chr20_-_29961589 | 0.66 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr20_+_31524009 | 0.65 |
|
|
|
| chr3_-_36298676 | 0.65 |
ENSDART00000157950
|
rogdi
|
rogdi homolog (Drosophila) |
| chr25_+_35742745 | 0.64 |
ENSDART00000152649
|
bmb
|
brambleberry |
| chr3_+_53913303 | 0.63 |
ENSDART00000109894
|
olfm2a
|
olfactomedin 2a |
| chr5_+_49724855 | 0.62 |
ENSDART00000167163
|
slf1
|
SMC5-SMC6 complex localization factor 1 |
| chr20_-_29961498 | 0.62 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr6_+_40525779 | 0.62 |
ENSDART00000033819
|
prkcda
|
protein kinase C, delta a |
| chr25_+_27300835 | 0.61 |
ENSDART00000103519
|
wasla
|
Wiskott-Aldrich syndrome-like a |
| chr12_+_21176669 | 0.61 |
ENSDART00000178562
|
ca10a
|
carbonic anhydrase Xa |
| chr4_+_9835529 | 0.61 |
ENSDART00000004879
|
hsp90b1
|
heat shock protein 90, beta (grp94), member 1 |
| chr20_-_31524684 | 0.60 |
ENSDART00000007735
|
ust
|
uronyl-2-sulfotransferase |
| chr16_+_39209567 | 0.59 |
ENSDART00000121756
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr16_+_23367580 | 0.59 |
ENSDART00000015956
|
efna1b
|
ephrin-A1b |
| chr2_-_20490037 | 0.58 |
ENSDART00000160388
|
FQ377605.1
|
ENSDARG00000101927 |
| chr17_+_24791024 | 0.58 |
ENSDART00000130871
|
spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr25_+_16259634 | 0.58 |
ENSDART00000136454
|
tead1a
|
TEA domain family member 1a |
| chr21_-_39132388 | 0.58 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr8_+_53674287 | 0.57 |
ENSDART00000166387
|
prickle2a
|
prickle homolog 2a |
| chr18_+_27593121 | 0.56 |
ENSDART00000134714
|
cd82b
|
CD82 molecule b |
| chr9_-_55395115 | 0.56 |
|
|
|
| chr1_-_44649941 | 0.56 |
ENSDART00000110390
|
ENSDARG00000025518
|
ENSDARG00000025518 |
| chr16_+_54579976 | 0.55 |
|
|
|
| chr19_-_9584480 | 0.55 |
ENSDART00000091615
|
iffo1a
|
intermediate filament family orphan 1a |
| chr6_-_11576632 | 0.53 |
ENSDART00000151717
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr22_-_11463487 | 0.53 |
ENSDART00000002691
|
tspan7b
|
tetraspanin 7b |
| chr18_+_17611571 | 0.53 |
ENSDART00000151850
|
herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr15_+_17164535 | 0.53 |
ENSDART00000155350
|
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr24_-_28227116 | 0.53 |
ENSDART00000148618
|
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr25_-_12538613 | 0.52 |
ENSDART00000162463
|
zcchc14
|
zinc finger, CCHC domain containing 14 |
| chr13_+_15685745 | 0.52 |
ENSDART00000137061
|
klc1a
|
kinesin light chain 1a |
| chr8_-_51612689 | 0.52 |
ENSDART00000175779
|
kctd9a
|
potassium channel tetramerization domain containing 9a |
| chr17_-_38940072 | 0.51 |
ENSDART00000141177
|
slc24a4a
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4a |
| chr19_-_19452999 | 0.51 |
ENSDART00000163359
ENSDART00000167951 |
dync1li1
|
dynein, cytoplasmic 1, light intermediate chain 1 |
| chr18_+_22804127 | 0.51 |
ENSDART00000149685
|
gnao1a
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a |
| chr24_+_39339095 | 0.51 |
ENSDART00000168705
|
si:ch73-103b11.2
|
si:ch73-103b11.2 |
| chr5_-_31173039 | 0.51 |
ENSDART00000137556
|
fam102ab
|
family with sequence similarity 102, member Ab |
| chr15_-_41277545 | 0.50 |
ENSDART00000157217
|
slc36a4
|
solute carrier family 36 (proton/amino acid symporter), member 4 |
| chr14_-_32937496 | 0.50 |
ENSDART00000048130
|
stard14
|
START domain containing 14 |
| chr22_+_32274241 | 0.50 |
ENSDART00000092082
|
manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr17_+_14957568 | 0.49 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| KN150226v1_+_11535 | 0.49 |
|
|
|
| chr1_+_30840599 | 0.49 |
ENSDART00000075286
|
slc2a15b
|
solute carrier family 2 (facilitated glucose transporter), member 15b |
| chr18_+_22804322 | 0.49 |
ENSDART00000149685
|
gnao1a
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a |
| chr2_-_15380727 | 0.48 |
|
|
|
| chr21_+_25031903 | 0.48 |
|
|
|
| chr7_+_66660882 | 0.48 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr6_-_13654186 | 0.48 |
ENSDART00000150102
ENSDART00000041269 |
cryba2a
|
crystallin, beta A2a |
| chr18_+_17611865 | 0.48 |
ENSDART00000141465
|
herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr8_+_15987710 | 0.47 |
ENSDART00000165141
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr8_+_40236654 | 0.47 |
ENSDART00000164414
|
orai1a
|
ORAI calcium release-activated calcium modulator 1a |
| chr25_-_17822003 | 0.47 |
ENSDART00000148660
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr12_+_19183576 | 0.47 |
ENSDART00000066391
|
csnk1e
|
casein kinase 1, epsilon |
| chr23_+_24685148 | 0.46 |
ENSDART00000134978
|
nckap5l
|
NCK-associated protein 5-like |
| chr18_+_22804023 | 0.46 |
ENSDART00000149685
|
gnao1a
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a |
| chr25_-_17822363 | 0.46 |
ENSDART00000023959
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr23_+_22954294 | 0.46 |
|
|
|
| chr19_+_20027309 | 0.46 |
ENSDART00000165934
|
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr5_-_47520995 | 0.45 |
ENSDART00000144252
|
BX465834.1
|
ENSDARG00000095715 |
| chr18_-_16134258 | 0.45 |
ENSDART00000061189
|
sspn
|
sarcospan (Kras oncogene-associated gene) |
| chr2_+_32033176 | 0.45 |
ENSDART00000135040
|
mycb
|
MYC proto-oncogene, bHLH transcription factor b |
| chr7_+_29930406 | 0.45 |
ENSDART00000075588
|
wdr76
|
WD repeat domain 76 |
| chr23_+_8862155 | 0.45 |
ENSDART00000132992
|
sox18
|
SRY (sex determining region Y)-box 18 |
| chr2_+_10023967 | 0.45 |
ENSDART00000148227
|
anxa13l
|
annexin A13, like |
| chr25_+_18613868 | 0.45 |
ENSDART00000011149
|
fam185a
|
family with sequence similarity 185, member A |
| chr22_-_29740737 | 0.44 |
ENSDART00000009223
|
pdcd4b
|
programmed cell death 4b |
| chr21_+_25031760 | 0.44 |
|
|
|
| chr11_+_43136945 | 0.44 |
|
|
|
| chr21_+_25032002 | 0.44 |
|
|
|
| chr5_+_33807197 | 0.44 |
ENSDART00000145127
|
lamc3
|
laminin, gamma 3 |
| chr7_+_38690837 | 0.43 |
ENSDART00000173565
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr6_+_27100249 | 0.43 |
ENSDART00000121558
|
atg4b
|
autophagy related 4B, cysteine peptidase |
| chr17_-_8481455 | 0.43 |
ENSDART00000148971
|
ctbp2a
|
C-terminal binding protein 2a |
| chr16_+_23976353 | 0.42 |
ENSDART00000058970
|
gtpbp10
|
GTP-binding protein 10 (putative) |
| chr15_-_19314712 | 0.42 |
ENSDART00000092705
|
igsf9ba
|
immunoglobulin superfamily, member 9Ba |
| chr13_-_39033893 | 0.42 |
ENSDART00000045434
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr5_-_31172791 | 0.42 |
ENSDART00000137556
|
fam102ab
|
family with sequence similarity 102, member Ab |
| chr4_-_20414881 | 0.42 |
ENSDART00000161343
|
cep83
|
centrosomal protein 83 |
| chr21_+_20439227 | 0.42 |
|
|
|
| chr10_-_34927807 | 0.42 |
ENSDART00000138755
|
dclk1a
|
doublecortin-like kinase 1a |
| chr5_-_66145078 | 0.41 |
ENSDART00000041441
|
stip1
|
stress-induced phosphoprotein 1 |
| chr13_-_42410700 | 0.41 |
ENSDART00000134533
|
btaf1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated |
| chr3_-_54352535 | 0.41 |
ENSDART00000021977
ENSDART00000078973 |
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr3_-_30730348 | 0.41 |
ENSDART00000154811
|
shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr21_-_21477462 | 0.40 |
ENSDART00000031205
|
pvrl3b
|
poliovirus receptor-related 3b |
| chr8_-_25547089 | 0.40 |
ENSDART00000078022
|
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr16_-_34331050 | 0.40 |
ENSDART00000081066
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr11_-_34909095 | 0.40 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr16_-_29502741 | 0.40 |
ENSDART00000148405
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr18_+_22804237 | 0.40 |
ENSDART00000149685
|
gnao1a
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a |
| chr13_-_42180169 | 0.40 |
ENSDART00000003706
|
kmo
|
kynurenine 3-monooxygenase |
| chr7_+_68964813 | 0.40 |
ENSDART00000166258
|
marveld3
|
MARVEL domain containing 3 |
| chr16_+_45370954 | 0.39 |
ENSDART00000147343
|
dnase2
|
deoxyribonuclease II, lysosomal |
| chr5_-_21015548 | 0.39 |
ENSDART00000040184
|
tenm1
|
teneurin transmembrane protein 1 |
| chr11_-_21143897 | 0.39 |
ENSDART00000163008
|
RASSF5
|
Ras association domain family member 5 |
| chr13_-_42598533 | 0.38 |
ENSDART00000160472
|
capn1a
|
calpain 1, (mu/I) large subunit a |
| chr5_-_36324311 | 0.38 |
ENSDART00000170733
|
kptn
|
kaptin (actin binding protein) |
| chr17_-_38939792 | 0.38 |
ENSDART00000141177
|
slc24a4a
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4a |
| chr22_+_18364282 | 0.38 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr19_-_44223619 | 0.38 |
ENSDART00000160879
|
klhl43
|
kelch-like family member 43 |
| chr25_-_17822194 | 0.38 |
ENSDART00000148660
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr16_-_39209426 | 0.38 |
|
|
|
| chr8_-_14014576 | 0.38 |
ENSDART00000135811
|
atp2b3a
|
ATPase, Ca++ transporting, plasma membrane 3a |
| chr2_-_45657620 | 0.38 |
ENSDART00000056357
|
gpsm2
|
G protein signaling modulator 2 |
| chr21_+_19958025 | 0.38 |
|
|
|
| chr16_-_51287698 | 0.38 |
ENSDART00000156255
|
ago1
|
argonaute RISC catalytic component 1 |
| chr2_+_49787853 | 0.37 |
ENSDART00000056254
|
stap2a
|
signal transducing adaptor family member 2a |
| chr6_-_31798839 | 0.37 |
|
|
|
| chr20_+_27040570 | 0.37 |
ENSDART00000138369
|
cdca4
|
cell division cycle associated 4 |
| chr16_+_4353561 | 0.37 |
ENSDART00000060729
|
yrdc
|
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr23_+_2481946 | 0.37 |
ENSDART00000126038
|
tcp1
|
t-complex 1 |
| chr18_+_17611931 | 0.37 |
ENSDART00000141465
|
herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr3_-_42985563 | 0.37 |
ENSDART00000176655
|
CU138533.3
|
ENSDARG00000108986 |
| chr11_+_40847576 | 0.37 |
ENSDART00000041531
|
park7
|
parkinson protein 7 |
| chr4_-_1871246 | 0.37 |
ENSDART00000103484
|
gas2l3
|
growth arrest-specific 2 like 3 |
| chr24_-_40968409 | 0.37 |
ENSDART00000169315
ENSDART00000171543 |
smyhc1
|
slow myosin heavy chain 1 |
| chr21_-_16123432 | 0.37 |
ENSDART00000124871
ENSDART00000161979 |
tnfrsfa
|
tumor necrosis factor receptor superfamily, member a |
| chr4_+_6634849 | 0.37 |
ENSDART00000099462
|
gpr85
|
G protein-coupled receptor 85 |
| chr4_-_24396003 | 0.36 |
|
|
|
| chr7_-_60046624 | 0.36 |
ENSDART00000127518
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr14_+_33382672 | 0.36 |
ENSDART00000075312
|
apln
|
apelin |
| chr22_-_111800 | 0.36 |
ENSDART00000163198
ENSDART00000168678 |
capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr6_+_19679720 | 0.36 |
ENSDART00000108946
|
lhfpl4a
|
lipoma HMGIC fusion partner-like 4a |
| chr5_+_43365449 | 0.36 |
ENSDART00000113502
|
TMEM8B
|
si:dkey-84j12.1 |
| chr10_-_4641624 | 0.36 |
ENSDART00000163951
|
plppr1
|
phospholipid phosphatase related 1 |
| chr16_+_53703721 | 0.36 |
ENSDART00000083534
|
gpatch3
|
G patch domain containing 3 |
| chr15_-_4589565 | 0.36 |
ENSDART00000008170
|
rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr15_-_35802739 | 0.36 |
ENSDART00000174714
|
CR848683.2
|
ENSDARG00000106192 |
| chr3_+_19471228 | 0.36 |
ENSDART00000025358
|
itgb3a
|
integrin beta 3a |
| chr2_-_48521151 | 0.35 |
ENSDART00000148788
|
per2
|
period circadian clock 2 |
| chr20_-_31524559 | 0.35 |
ENSDART00000007735
|
ust
|
uronyl-2-sulfotransferase |
| chr11_-_42022975 | 0.35 |
|
|
|
| chr18_+_18598885 | 0.35 |
|
|
|
| chr14_-_6939857 | 0.35 |
ENSDART00000108796
|
stox2b
|
storkhead box 2b |
| chr9_+_29737843 | 0.35 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr6_+_27100350 | 0.35 |
ENSDART00000121558
|
atg4b
|
autophagy related 4B, cysteine peptidase |
| chr12_+_22137718 | 0.35 |
ENSDART00000131175
|
wnt3
|
wingless-type MMTV integration site family, member 3 |
| chr13_-_36164510 | 0.35 |
ENSDART00000169768
|
slc8a3
|
solute carrier family 8 (sodium/calcium exchanger), member 3 |
| chr12_-_45496246 | 0.35 |
ENSDART00000176229
ENSDART00000175457 |
FO818685.1
|
ENSDARG00000107895 |
| chr10_-_4641512 | 0.35 |
ENSDART00000163951
|
plppr1
|
phospholipid phosphatase related 1 |
| chr24_+_17190448 | 0.35 |
ENSDART00000159724
ENSDART00000081851 ENSDART00000018868 |
pdia4
|
protein disulfide isomerase family A, member 4 |
| chr20_-_29961621 | 0.35 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr7_-_60046465 | 0.35 |
ENSDART00000127518
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr14_+_20595818 | 0.35 |
ENSDART00000079452
|
lygl1
|
lysozyme g-like 1 |
| chr13_-_18559962 | 0.34 |
ENSDART00000057867
|
sfxn3
|
sideroflexin 3 |
| chr4_+_9278724 | 0.34 |
ENSDART00000048707
|
srgap1b
|
SLIT-ROBO Rho GTPase activating protein 1b |
| chr1_-_55072271 | 0.34 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr15_+_19717045 | 0.34 |
ENSDART00000138680
|
lim2.3
|
lens intrinsic membrane protein 2.3 |
| chr5_-_68408107 | 0.34 |
ENSDART00000109761
ENSDART00000156681 ENSDART00000167680 |
ENSDARG00000076888
|
ENSDARG00000076888 |
| chr19_+_2942485 | 0.34 |
ENSDART00000177848
|
CABZ01066434.1
|
ENSDARG00000107451 |
| chr9_-_34691081 | 0.34 |
ENSDART00000114043
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr8_+_40176716 | 0.34 |
ENSDART00000055861
|
rnf34a
|
ring finger protein 34a |
| chr3_-_20832951 | 0.34 |
ENSDART00000160794
|
slc35b1
|
solute carrier family 35, member B1 |
| chr20_+_340283 | 0.34 |
ENSDART00000036635
|
fynb
|
FYN proto-oncogene, Src family tyrosine kinase b |
| chr1_+_45907269 | 0.34 |
ENSDART00000132861
|
cab39l
|
calcium binding protein 39-like |
| chr15_-_43283562 | 0.34 |
ENSDART00000110352
|
ap1s3a
|
adaptor-related protein complex 1, sigma 3 subunit, a |
| chr8_-_16638825 | 0.33 |
ENSDART00000100727
|
osbpl9
|
oxysterol binding protein-like 9 |
| chr4_-_17364090 | 0.33 |
ENSDART00000134467
|
parpbp
|
PARP1 binding protein |
| chr20_-_8431338 | 0.33 |
ENSDART00000145841
ENSDART00000143936 ENSDART00000083906 ENSDART00000167102 ENSDART00000083908 |
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr12_+_7421078 | 0.33 |
ENSDART00000163114
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr18_-_616875 | 0.33 |
ENSDART00000169974
|
sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr13_-_23626146 | 0.33 |
ENSDART00000057612
|
rgs17
|
regulator of G protein signaling 17 |
| chr22_-_29740287 | 0.33 |
ENSDART00000166002
|
pdcd4b
|
programmed cell death 4b |
| chr4_-_20514831 | 0.33 |
ENSDART00000146621
|
stk38l
|
serine/threonine kinase 38 like |
| chr3_+_59972297 | 0.33 |
|
|
|
| chr6_-_53145759 | 0.33 |
ENSDART00000154429
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr12_+_21176777 | 0.33 |
ENSDART00000178562
|
ca10a
|
carbonic anhydrase Xa |
| chr14_-_32063302 | 0.33 |
ENSDART00000173114
|
fgf13a
|
fibroblast growth factor 13a |
| chr16_+_41222408 | 0.32 |
ENSDART00000128963
|
nek11
|
NIMA-related kinase 11 |
| chr2_-_32754956 | 0.32 |
ENSDART00000135293
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr18_+_20058174 | 0.32 |
ENSDART00000146957
|
uacaa
|
uveal autoantigen with coiled-coil domains and ankyrin repeats a |
| chr1_-_44892600 | 0.32 |
ENSDART00000149155
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr8_-_51612596 | 0.32 |
ENSDART00000175779
|
kctd9a
|
potassium channel tetramerization domain containing 9a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:1902235 | regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902235) negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.3 | 0.9 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) negative regulation of calcium ion import(GO:0090281) |
| 0.3 | 0.8 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.2 | 0.9 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 0.6 | GO:0007344 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.2 | 0.7 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.2 | 0.7 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.2 | 0.5 | GO:0042128 | nitrate metabolic process(GO:0042126) nitrate assimilation(GO:0042128) |
| 0.2 | 0.5 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.1 | 0.6 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.1 | 0.4 | GO:0043420 | anthranilate metabolic process(GO:0043420) |
| 0.1 | 0.4 | GO:0039535 | negative regulation of type I interferon production(GO:0032480) cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) cellular response to virus(GO:0098586) |
| 0.1 | 0.4 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.1 | 0.4 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) |
| 0.1 | 0.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.3 | GO:0044036 | cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.1 | 0.5 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.8 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.1 | 0.4 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 0.5 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.3 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 0.6 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.1 | 0.4 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 0.5 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.8 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 0.5 | GO:0072584 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.3 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.5 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.2 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 1.8 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 0.3 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.1 | 0.2 | GO:1903533 | regulation of protein targeting(GO:1903533) |
| 0.1 | 0.5 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.4 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 0.7 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.1 | 0.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.5 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.4 | GO:0032371 | regulation of sterol transport(GO:0032371) regulation of cholesterol transport(GO:0032374) |
| 0.1 | 2.6 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.1 | 0.4 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.3 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.3 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.1 | 1.1 | GO:0001840 | neural plate development(GO:0001840) |
| 0.1 | 0.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.2 | GO:2000623 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 0.6 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.6 | GO:0061647 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.1 | 0.3 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.4 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.1 | 0.6 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.0 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.1 | 2.1 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.5 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.4 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 0.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 1.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.2 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.1 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.8 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.6 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.7 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.4 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.3 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.4 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.9 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.0 | 0.4 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.4 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.3 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.2 | GO:0019229 | regulation of vasoconstriction(GO:0019229) |
| 0.0 | 0.3 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.3 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.4 | GO:1903432 | regulation of TORC1 signaling(GO:1903432) |
| 0.0 | 0.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.9 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0002762 | negative regulation of myeloid leukocyte differentiation(GO:0002762) regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.3 | GO:0046037 | GMP biosynthetic process(GO:0006177) GTP biosynthetic process(GO:0006183) GMP metabolic process(GO:0046037) |
| 0.0 | 0.1 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.0 | 0.1 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.5 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.0 | 0.4 | GO:0010632 | regulation of epithelial cell migration(GO:0010632) |
| 0.0 | 0.1 | GO:1902871 | negative regulation of photoreceptor cell differentiation(GO:0046533) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 1.0 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.2 | GO:1904666 | regulation of ubiquitin protein ligase activity(GO:1904666) |
| 0.0 | 0.2 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.2 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.3 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.4 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.0 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.3 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.2 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 0.1 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.3 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.2 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.5 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.2 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.4 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 1.2 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.2 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.2 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.4 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.3 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.7 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.2 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.0 | 0.3 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.2 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.7 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.7 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.3 | GO:0032456 | endocytic recycling(GO:0032456) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.2 | 0.6 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 0.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.5 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 1.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.9 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 2.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.2 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.1 | 0.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.8 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.8 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.2 | 1.9 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 0.7 | GO:0005460 | UDP-glucose transmembrane transporter activity(GO:0005460) |
| 0.2 | 0.5 | GO:0008942 | nitrite reductase [NAD(P)H] activity(GO:0008942) oxidoreductase activity, acting on other nitrogenous compounds as donors, with NAD or NADP as acceptor(GO:0046857) |
| 0.2 | 0.9 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.4 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.1 | 0.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.8 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.5 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.3 | GO:0004617 | phosphoglycerate dehydrogenase activity(GO:0004617) |
| 0.1 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 1.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.5 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.1 | 0.6 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.1 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.6 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.1 | 1.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 1.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.2 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.1 | 0.2 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 1.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.2 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.1 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.3 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 0.2 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 1.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.7 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.5 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.3 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.6 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.6 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.0 | 0.1 | GO:0035256 | glutamate receptor binding(GO:0035254) G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.5 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.5 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 1.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.9 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 1.1 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.2 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 0.8 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.8 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.8 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.4 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 1.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 0.9 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 0.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 0.9 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 0.9 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 0.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 0.5 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 1.5 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 0.5 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.9 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.3 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.2 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.9 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |