Project
DANIO-CODE
Navigation
Downloads
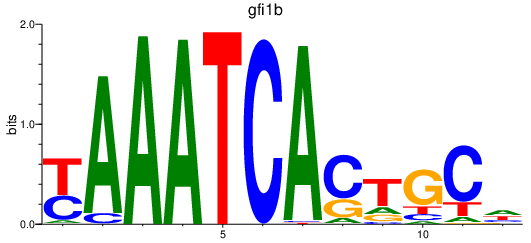
Results for gfi1b
Z-value: 0.90
Transcription factors associated with gfi1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gfi1b
|
ENSDARG00000079947 | growth factor independent 1B transcription repressor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gfi1b | dr10_dc_chr21_-_17259886_17259933 | 0.88 | 5.9e-06 | Click! |
Activity profile of gfi1b motif
Sorted Z-values of gfi1b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of gfi1b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_43039606 | 3.65 |
ENSDART00000010104
|
crtap
|
cartilage associated protein |
| chr3_-_32686790 | 3.13 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr23_-_35595271 | 3.13 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr17_+_15425559 | 2.94 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr7_-_25623974 | 2.70 |
ENSDART00000173602
|
cd99l2
|
CD99 molecule-like 2 |
| chr5_-_39910235 | 2.61 |
ENSDART00000146237
ENSDART00000163302 |
fsta
|
follistatin a |
| chr16_-_45269179 | 2.52 |
ENSDART00000162095
|
si:dkey-33i11.4
|
si:dkey-33i11.4 |
| chr15_-_24987099 | 2.24 |
|
|
|
| chr6_+_29800606 | 2.14 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr2_+_32036450 | 2.08 |
ENSDART00000140776
|
CR391940.1
|
ENSDARG00000091946 |
| chr25_-_31881585 | 1.89 |
ENSDART00000123999
ENSDART00000158091 |
ENSDARG00000100731
|
ENSDARG00000100731 |
| chr14_+_34683602 | 1.83 |
ENSDART00000172171
|
ebf1a
|
early B-cell factor 1a |
| chr8_-_19364689 | 1.80 |
ENSDART00000014183
|
colgalt2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr23_+_8671759 | 1.73 |
ENSDART00000129997
|
rgs19
|
regulator of G protein signaling 19 |
| chr19_-_28546417 | 1.69 |
ENSDART00000130922
ENSDART00000079114 |
irx1b
|
iroquois homeobox 1b |
| chr6_+_7309023 | 1.62 |
ENSDART00000160128
|
erbb3a
|
erb-b2 receptor tyrosine kinase 3a |
| chr1_+_13779755 | 1.59 |
|
|
|
| chr1_+_12076622 | 1.56 |
ENSDART00000159226
|
pcdh10a
|
protocadherin 10a |
| chr12_-_3742062 | 1.52 |
ENSDART00000092983
|
ENSDARG00000063555
|
ENSDARG00000063555 |
| chr2_-_10555152 | 1.51 |
ENSDART00000150166
|
gng12a
|
guanine nucleotide binding protein (G protein), gamma 12a |
| chr8_-_15144533 | 1.49 |
ENSDART00000138855
|
bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr4_-_12008472 | 1.48 |
ENSDART00000092250
|
btbd11a
|
BTB (POZ) domain containing 11a |
| chr9_+_48309684 | 1.46 |
ENSDART00000177287
ENSDART00000113883 |
ENSDARG00000041339
|
ENSDARG00000041339 |
| chr23_-_10202347 | 1.45 |
ENSDART00000142442
|
plxnb1a
|
plexin b1a |
| chr23_+_23305483 | 1.40 |
ENSDART00000126479
|
plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| KN150342v1_-_59821 | 1.40 |
ENSDART00000163274
|
ENSDARG00000100757
|
ENSDARG00000100757 |
| chr7_-_70219217 | 1.36 |
ENSDART00000097710
|
ppargc1a
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr7_+_25649559 | 1.35 |
ENSDART00000026295
|
arrb2b
|
arrestin, beta 2b |
| chr1_-_24988461 | 1.33 |
ENSDART00000054230
|
fgg
|
fibrinogen gamma chain |
| chr20_+_51034244 | 1.32 |
|
|
|
| chr3_+_23587156 | 1.31 |
|
|
|
| chr7_+_20699191 | 1.26 |
ENSDART00000062003
|
efnb3b
|
ephrin-B3b |
| chr18_+_23891068 | 1.25 |
ENSDART00000146490
|
BX569787.1
|
ENSDARG00000092402 |
| chr10_-_39210658 | 1.22 |
ENSDART00000150193
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr6_-_24003717 | 1.20 |
ENSDART00000164915
|
scinla
|
scinderin like a |
| chr24_-_4418454 | 1.19 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr10_+_9325719 | 1.19 |
ENSDART00000064968
|
rasgef1bb
|
RasGEF domain family, member 1Bb |
| chr6_-_12616971 | 1.18 |
ENSDART00000056764
|
bmpr2a
|
bone morphogenetic protein receptor, type II a (serine/threonine kinase) |
| chr20_+_19175518 | 1.18 |
|
|
|
| chr20_-_51746555 | 1.16 |
ENSDART00000065231
|
disp1
|
dispatched homolog 1 (Drosophila) |
| chr21_-_17566461 | 1.14 |
ENSDART00000044078
ENSDART00000168495 ENSDART00000020048 |
gsna
|
gelsolin a |
| chr13_+_21649283 | 1.13 |
ENSDART00000021556
|
si:ch211-51a6.2
|
si:ch211-51a6.2 |
| chr25_+_11317025 | 1.09 |
|
|
|
| chr5_-_58162552 | 1.08 |
ENSDART00000161230
|
arhgef12b
|
Rho guanine nucleotide exchange factor (GEF) 12b |
| chr25_+_19901741 | 1.06 |
ENSDART00000026401
|
ENSDARG00000004577
|
ENSDARG00000004577 |
| chr7_+_48047139 | 1.04 |
ENSDART00000052122
|
crabp1b
|
cellular retinoic acid binding protein 1b |
| chr23_-_10202175 | 1.04 |
ENSDART00000142442
|
plxnb1a
|
plexin b1a |
| chr14_+_34683695 | 1.03 |
ENSDART00000170631
|
ebf1a
|
early B-cell factor 1a |
| chr6_+_35378839 | 1.02 |
ENSDART00000102483
ENSDART00000133783 |
rgs4
|
regulator of G protein signaling 4 |
| chr9_+_21466934 | 1.01 |
ENSDART00000139620
|
lats2
|
large tumor suppressor kinase 2 |
| chr12_-_3741925 | 1.00 |
ENSDART00000092983
|
ENSDARG00000063555
|
ENSDARG00000063555 |
| chr19_-_22904687 | 0.98 |
ENSDART00000141503
|
pleca
|
plectin a |
| chr3_-_32409303 | 0.98 |
ENSDART00000151476
|
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr23_-_20494890 | 0.92 |
|
|
|
| chr8_+_2260519 | 0.91 |
ENSDART00000136743
|
ikbkb
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
| chr9_-_5618609 | 0.91 |
ENSDART00000127219
|
FAM155A
|
family with sequence similarity 155 member A |
| chr23_+_7537089 | 0.91 |
ENSDART00000135551
|
si:ch211-200e2.1
|
si:ch211-200e2.1 |
| chr18_+_23891152 | 0.90 |
ENSDART00000146490
|
BX569787.1
|
ENSDARG00000092402 |
| chr23_+_14058972 | 0.90 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr2_+_10023967 | 0.90 |
ENSDART00000148227
|
anxa13l
|
annexin A13, like |
| chr22_-_36552513 | 0.90 |
ENSDART00000129318
|
CABZ01045212.1
|
ENSDARG00000087525 |
| chr3_+_22246697 | 0.89 |
ENSDART00000155597
|
arhgap27l
|
Rho GTPase activating protein 27, like |
| chr24_+_10276926 | 0.89 |
ENSDART00000145771
ENSDART00000157350 |
CT573344.1
|
ENSDARG00000093108 |
| chr13_-_43027245 | 0.88 |
ENSDART00000099601
|
ENSDARG00000068784
|
ENSDARG00000068784 |
| chr9_-_1964814 | 0.88 |
ENSDART00000082354
|
hoxd9a
|
homeobox D9a |
| chr3_+_23561215 | 0.87 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr1_-_46398498 | 0.87 |
ENSDART00000000087
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr19_+_47846675 | 0.85 |
ENSDART00000041114
|
psmb2
|
proteasome subunit beta 2 |
| chr16_+_10531986 | 0.85 |
ENSDART00000161568
|
ino80e
|
INO80 complex subunit E |
| chr11_+_34788205 | 0.85 |
ENSDART00000124800
|
fam212aa
|
family with sequence similarity 212, member Aa |
| chr16_+_10531926 | 0.85 |
ENSDART00000161568
|
ino80e
|
INO80 complex subunit E |
| chr14_-_29565869 | 0.84 |
ENSDART00000172349
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr21_+_38264846 | 0.83 |
ENSDART00000065159
|
zgc:158291
|
zgc:158291 |
| chr13_+_21649237 | 0.83 |
ENSDART00000021556
|
si:ch211-51a6.2
|
si:ch211-51a6.2 |
| chr24_+_9272045 | 0.82 |
ENSDART00000132724
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr2_+_2583068 | 0.82 |
|
|
|
| chr7_-_26137243 | 0.81 |
ENSDART00000111494
|
gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr21_-_33961137 | 0.80 |
ENSDART00000100508
|
ebf1b
|
early B-cell factor 1b |
| chr20_+_27565701 | 0.79 |
|
|
|
| chr19_-_35642440 | 0.78 |
ENSDART00000167853
ENSDART00000054274 |
macf1a
|
microtubule-actin crosslinking factor 1a |
| chr2_-_32468715 | 0.74 |
|
|
|
| chr7_+_44106327 | 0.74 |
ENSDART00000108766
ENSDART00000111441 |
cdh5
|
cadherin 5 |
| chr19_+_2291874 | 0.72 |
|
|
|
| chr20_+_18263793 | 0.72 |
ENSDART00000011287
|
aqp4
|
aquaporin 4 |
| chr7_-_57727148 | 0.71 |
|
|
|
| chr3_+_23561502 | 0.70 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr9_+_34832049 | 0.70 |
ENSDART00000100735
ENSDART00000133996 |
shox
|
short stature homeobox |
| chr8_+_39525254 | 0.70 |
|
|
|
| chr7_+_48047113 | 0.68 |
ENSDART00000052122
|
crabp1b
|
cellular retinoic acid binding protein 1b |
| chr23_+_22729939 | 0.68 |
ENSDART00000009337
|
eno1a
|
enolase 1a, (alpha) |
| chr6_-_16156494 | 0.67 |
|
|
|
| chr18_-_22745846 | 0.64 |
ENSDART00000023721
|
nudt21
|
nudix hydrolase 21 |
| chr18_+_23890892 | 0.64 |
ENSDART00000146490
|
BX569787.1
|
ENSDARG00000092402 |
| chr3_-_12818954 | 0.63 |
ENSDART00000158747
ENSDART00000158815 |
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr17_-_12231165 | 0.63 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr6_+_45493396 | 0.62 |
ENSDART00000159863
|
cntn4
|
contactin 4 |
| chr25_-_14953284 | 0.61 |
ENSDART00000165632
|
pax6a
|
paired box 6a |
| chr14_-_9407573 | 0.61 |
ENSDART00000166739
|
NRK
|
si:zfos-2326c3.2 |
| KN150342v1_-_60050 | 0.60 |
ENSDART00000163274
|
ENSDARG00000100757
|
ENSDARG00000100757 |
| chr22_+_31866783 | 0.60 |
ENSDART00000159825
|
dock3
|
dedicator of cytokinesis 3 |
| chr6_-_34237257 | 0.60 |
ENSDART00000102391
|
dmrt2b
|
doublesex and mab-3 related transcription factor 2b |
| chr24_+_35500964 | 0.59 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr12_+_38388606 | 0.59 |
ENSDART00000081046
|
ttyh2
|
tweety family member 2 |
| chr12_+_5969088 | 0.56 |
ENSDART00000114961
|
si:ch211-131k2.3
|
si:ch211-131k2.3 |
| chr7_-_25624128 | 0.54 |
ENSDART00000173505
|
cd99l2
|
CD99 molecule-like 2 |
| chr10_+_9134634 | 0.54 |
ENSDART00000110443
|
fstb
|
follistatin b |
| chr9_+_3048953 | 0.54 |
ENSDART00000140614
|
BX957292.1
|
ENSDARG00000092185 |
| chr23_+_35980812 | 0.52 |
|
|
|
| chr20_-_26142897 | 0.52 |
ENSDART00000140255
|
si:dkey-12h9.6
|
si:dkey-12h9.6 |
| chr16_+_23367625 | 0.51 |
ENSDART00000015956
|
efna1b
|
ephrin-A1b |
| chr3_+_23561306 | 0.50 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr3_+_29891683 | 0.48 |
|
|
|
| chr21_-_16123432 | 0.46 |
ENSDART00000124871
ENSDART00000161979 |
tnfrsfa
|
tumor necrosis factor receptor superfamily, member a |
| chr12_+_27444832 | 0.44 |
ENSDART00000133719
|
etv4
|
ets variant 4 |
| chr2_+_56705456 | 0.44 |
|
|
|
| chr6_-_23830912 | 0.42 |
|
|
|
| chr6_+_50393779 | 0.42 |
ENSDART00000055502
|
ergic3
|
ERGIC and golgi 3 |
| chr5_+_69401770 | 0.42 |
|
|
|
| chr12_-_3742020 | 0.41 |
ENSDART00000092983
|
ENSDARG00000063555
|
ENSDARG00000063555 |
| chr25_-_3853819 | 0.40 |
ENSDART00000124749
|
myrf
|
myelin regulatory factor |
| chr9_+_50609926 | 0.38 |
ENSDART00000098687
|
GRB14
|
growth factor receptor bound protein 14 |
| chr6_+_11014565 | 0.35 |
ENSDART00000132677
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr5_+_57102973 | 0.32 |
ENSDART00000135344
|
ppp2r1ba
|
protein phosphatase 2, regulatory subunit A, beta a |
| chr7_+_48047266 | 0.32 |
ENSDART00000052122
|
crabp1b
|
cellular retinoic acid binding protein 1b |
| chr17_-_37447917 | 0.31 |
ENSDART00000075975
|
crip1
|
cysteine-rich protein 1 |
| chr23_-_24336930 | 0.31 |
ENSDART00000160312
|
hspb7
|
heat shock protein family, member 7 (cardiovascular) |
| chr6_+_3356712 | 0.30 |
ENSDART00000151607
|
faah
|
fatty acid amide hydrolase |
| chr5_-_11593397 | 0.30 |
ENSDART00000161542
|
lrrc74b
|
leucine rich repeat containing 74B |
| chr3_-_30810423 | 0.30 |
ENSDART00000076830
|
coro1a
|
coronin, actin binding protein, 1A |
| chr7_-_27877036 | 0.30 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr6_+_3356617 | 0.30 |
ENSDART00000109576
|
faah
|
fatty acid amide hydrolase |
| chr9_+_32267297 | 0.29 |
|
|
|
| chr8_-_25678085 | 0.29 |
ENSDART00000143938
|
sema3ga
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ga |
| chr8_-_16479344 | 0.27 |
ENSDART00000146469
ENSDART00000132681 |
ttc39a
|
tetratricopeptide repeat domain 39A |
| chr15_+_32863182 | 0.25 |
|
|
|
| chr25_-_14953370 | 0.24 |
ENSDART00000159490
|
pax6a
|
paired box 6a |
| chr20_-_28226 | 0.24 |
ENSDART00000148717
|
zgc:174972
|
zgc:174972 |
| chr14_-_46525518 | 0.21 |
ENSDART00000173172
|
si:ch211-114c17.1
|
si:ch211-114c17.1 |
| chr9_+_44921233 | 0.20 |
ENSDART00000145271
|
nckap1
|
NCK-associated protein 1 |
| chr2_+_31855153 | 0.19 |
ENSDART00000066789
|
stard3nl
|
STARD3 N-terminal like |
| chr19_+_20617339 | 0.19 |
ENSDART00000133633
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr19_+_20616655 | 0.18 |
ENSDART00000163026
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr3_-_30833046 | 0.18 |
|
|
|
| chr10_-_34927807 | 0.16 |
ENSDART00000138755
|
dclk1a
|
doublecortin-like kinase 1a |
| chr25_+_17906515 | 0.16 |
|
|
|
| KN150529v1_+_16214 | 0.15 |
ENSDART00000177459
|
CABZ01084538.1
|
ENSDARG00000108298 |
| chr3_-_41944723 | 0.14 |
ENSDART00000083111
|
ttyh3a
|
tweety family member 3a |
| chr12_-_4183910 | 0.14 |
ENSDART00000129427
|
vkorc1
|
vitamin K epoxide reductase complex, subunit 1 |
| chr21_+_27333941 | 0.14 |
ENSDART00000009234
|
rbm14a
|
RNA binding motif protein 14a |
| chr23_+_25930072 | 0.12 |
ENSDART00000124103
|
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr9_+_19678618 | 0.11 |
ENSDART00000152034
|
si:ch211-140m22.7
|
si:ch211-140m22.7 |
| chr23_+_8862155 | 0.09 |
ENSDART00000132992
|
sox18
|
SRY (sex determining region Y)-box 18 |
| chr16_+_10438774 | 0.09 |
ENSDART00000121966
|
mdc1
|
mediator of DNA damage checkpoint 1 |
| chr17_-_19809993 | 0.08 |
|
|
|
| chr10_-_35598016 | 0.06 |
ENSDART00000162139
|
si:ch211-244c8.4
|
si:ch211-244c8.4 |
| chr4_-_12796590 | 0.05 |
ENSDART00000075127
|
b2m
|
beta-2-microglobulin |
| chr25_+_17217050 | 0.05 |
ENSDART00000125459
|
cnot1
|
CCR4-NOT transcription complex, subunit 1 |
| chr25_-_34647197 | 0.02 |
ENSDART00000114908
|
si:dkey-261m9.15
|
si:dkey-261m9.15 |
| chr25_+_35791595 | 0.02 |
ENSDART00000103013
|
ENSDARG00000070286
|
ENSDARG00000070286 |
| chr6_-_16156294 | 0.02 |
ENSDART00000160809
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0072025 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.5 | 1.4 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) regulation of protein binding(GO:0043393) |
| 0.4 | 3.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.4 | 1.2 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.4 | 2.5 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.3 | 0.8 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.3 | 2.0 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.2 | 0.7 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.2 | 3.6 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.2 | 0.7 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.2 | 1.6 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 0.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 1.5 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 0.9 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 1.3 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 1.0 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.1 | 0.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.5 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 0.4 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 0.7 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 2.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.4 | GO:0043217 | myelin maintenance(GO:0043217) |
| 0.1 | 0.8 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.1 | 0.9 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.6 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.6 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.3 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.1 | 1.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.2 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 1.0 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.0 | 0.3 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.9 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 2.0 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 1.2 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.0 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.7 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.6 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 0.1 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 1.7 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.3 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.6 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.0 | 0.7 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 1.7 | GO:0030902 | hindbrain development(GO:0030902) |
| 0.0 | 1.3 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.4 | 2.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.2 | 0.8 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.2 | 0.9 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 0.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 1.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.5 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 1.7 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 1.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.9 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.6 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.9 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 1.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 1.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.8 | GO:0005925 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.4 | 1.8 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.3 | 0.9 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.3 | 3.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.3 | 2.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 0.8 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.2 | 2.0 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 3.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 0.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 2.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 0.6 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.1 | 0.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 1.0 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.1 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.1 | 2.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 3.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 2.0 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.9 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.6 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.6 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 4.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.0 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 1.2 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.4 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 5.6 | GO:0005509 | calcium ion binding(GO:0005509) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 0.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 0.7 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.6 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 3.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 0.9 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 3.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.7 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 0.7 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 1.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |