Project
DANIO-CODE
Navigation
Downloads
Results for gli1+gli2b
Z-value: 1.14
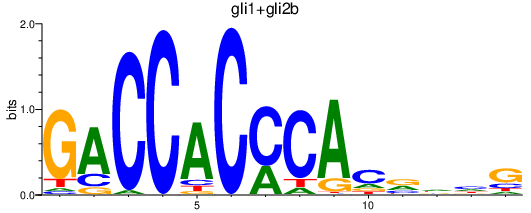
Transcription factors associated with gli1+gli2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gli2b
|
ENSDARG00000020884 | GLI family zinc finger 2b |
|
gli1
|
ENSDARG00000101244 | GLI family zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gli1 | dr10_dc_chr6_-_59510397_59510468 | 0.92 | 4.7e-07 | Click! |
Activity profile of gli1+gli2b motif
Sorted Z-values of gli1+gli2b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of gli1+gli2b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_20496283 | 3.83 |
ENSDART00000144047
ENSDART00000104336 |
tnnc2
|
troponin C type 2 (fast) |
| chr7_+_29680967 | 3.72 |
ENSDART00000173540
|
tpma
|
alpha-tropomyosin |
| chr8_+_29953399 | 3.41 |
ENSDART00000149372
ENSDART00000007640 |
ptch1
ptch1
|
patched 1 patched 1 |
| chr20_-_54206610 | 3.27 |
ENSDART00000004756
|
hsp90aa1.1
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 1 |
| chr2_-_34010299 | 3.09 |
ENSDART00000140910
|
ptch2
|
patched 2 |
| chr18_-_5796081 | 2.89 |
ENSDART00000097696
ENSDART00000151453 |
si:ch1073-459b3.2
|
si:ch1073-459b3.2 |
| chr23_-_22204223 | 2.67 |
ENSDART00000079212
|
phc2a
|
polyhomeotic homolog 2a (Drosophila) |
| chr7_-_26767182 | 2.67 |
ENSDART00000052731
|
nucb2a
|
nucleobindin 2a |
| chr10_+_43360137 | 2.59 |
ENSDART00000012522
|
vcanb
|
versican b |
| chr5_-_28090077 | 2.59 |
|
|
|
| chr6_+_6967174 | 2.48 |
ENSDART00000151311
|
ENSDARG00000058903
|
ENSDARG00000058903 |
| chr10_+_33227967 | 2.44 |
ENSDART00000159666
|
myl10
|
myosin, light chain 10, regulatory |
| chr11_-_80746 | 2.32 |
|
|
|
| chr23_-_27226280 | 2.30 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr6_-_22504772 | 2.27 |
ENSDART00000170039
|
sept9b
|
septin 9b |
| chr10_-_24422257 | 2.26 |
ENSDART00000141332
ENSDART00000100772 |
slc43a2b
|
solute carrier family 43 (amino acid system L transporter), member 2b |
| chr21_-_43020159 | 2.23 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr4_+_10722966 | 2.17 |
ENSDART00000102534
ENSDART00000067253 ENSDART00000136000 |
stab2
|
stabilin 2 |
| chr23_+_34079219 | 1.94 |
ENSDART00000132668
|
si:ch211-207e14.4
|
si:ch211-207e14.4 |
| chr9_-_23081250 | 1.92 |
|
|
|
| chr12_-_17357133 | 1.92 |
ENSDART00000079115
|
papss2b
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2b |
| chr10_-_14530112 | 1.81 |
ENSDART00000176126
|
galt
|
galactose-1-phosphate uridylyltransferase |
| chr8_+_13064944 | 1.76 |
|
|
|
| chr23_+_28656263 | 1.75 |
ENSDART00000020296
|
nadl1.2
|
neural adhesion molecule L1.2 |
| chr12_+_27026112 | 1.72 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr13_-_39821399 | 1.70 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr8_-_40287789 | 1.69 |
ENSDART00000074125
|
aplnra
|
apelin receptor a |
| chr12_-_457997 | 1.69 |
ENSDART00000143232
|
dhrs7cb
|
dehydrogenase/reductase (SDR family) member 7Cb |
| chr21_-_30378764 | 1.67 |
ENSDART00000136247
|
grpel2
|
GrpE-like 2, mitochondrial |
| chr23_+_4475256 | 1.67 |
ENSDART00000081821
|
wnt7ab
|
wingless-type MMTV integration site family, member 7Ab |
| chr24_-_2278409 | 1.63 |
|
|
|
| chr15_-_31687 | 1.61 |
|
|
|
| chr8_+_999710 | 1.59 |
ENSDART00000081432
|
sprb
|
sepiapterin reductase b |
| chr23_-_27226387 | 1.59 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr22_+_24362712 | 1.58 |
ENSDART00000157861
|
p3h2
|
prolyl 3-hydroxylase 2 |
| chr24_-_16884946 | 1.56 |
ENSDART00000171988
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr15_-_42250788 | 1.55 |
ENSDART00000015843
|
pax3b
|
paired box 3b |
| chr20_-_28739099 | 1.51 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr12_+_27370834 | 1.49 |
ENSDART00000105661
|
meox1
|
mesenchyme homeobox 1 |
| chr21_-_91068 | 1.47 |
|
|
|
| chr14_-_16171165 | 1.47 |
ENSDART00000089021
|
canx
|
calnexin |
| chr9_-_10561062 | 1.47 |
ENSDART00000175269
|
thsd7ba
|
thrombospondin, type I, domain containing 7Ba |
| chr8_-_4903896 | 1.47 |
ENSDART00000004986
|
igsf9b
|
immunoglobulin superfamily, member 9b |
| chr4_+_5124186 | 1.46 |
ENSDART00000127874
|
ccnd2b
|
cyclin D2, b |
| chr21_-_37404394 | 1.46 |
ENSDART00000129439
|
BX649250.1
|
ENSDARG00000091187 |
| chr9_-_46272322 | 1.43 |
ENSDART00000169682
|
hdac4
|
histone deacetylase 4 |
| chr6_+_9192009 | 1.43 |
ENSDART00000022620
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr13_-_9554501 | 1.41 |
ENSDART00000139541
|
ENSDARG00000069828
|
ENSDARG00000069828 |
| chr3_-_15918262 | 1.41 |
ENSDART00000157315
|
ENSDARG00000097322
|
ENSDARG00000097322 |
| chr19_-_7069920 | 1.40 |
ENSDART00000145741
|
znf384l
|
zinc finger protein 384 like |
| chr6_+_45930323 | 1.37 |
ENSDART00000154896
|
rbp7b
|
retinol binding protein 7b, cellular |
| chr23_-_20402258 | 1.35 |
ENSDART00000136204
|
CR749762.2
|
ENSDARG00000093223 |
| chr15_-_31871 | 1.25 |
|
|
|
| chr4_+_27141061 | 1.25 |
ENSDART00000145083
|
brd1a
|
bromodomain containing 1a |
| chr13_-_40628582 | 1.18 |
ENSDART00000111641
ENSDART00000159255 |
morn4
|
MORN repeat containing 4 |
| chr2_-_49114158 | 1.14 |
|
|
|
| chr2_+_47885551 | 1.11 |
|
|
|
| chr16_+_12740993 | 1.09 |
ENSDART00000128497
|
nat14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr19_-_12785952 | 1.03 |
|
|
|
| chr5_+_9542042 | 1.00 |
ENSDART00000137543
|
gstt2
|
glutathione S-transferase theta 2 |
| chr3_+_47412661 | 0.96 |
ENSDART00000155648
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr17_-_33764165 | 0.95 |
ENSDART00000043651
|
dnal1
|
dynein, axonemal, light chain 1 |
| chr13_-_40628537 | 0.93 |
ENSDART00000111641
ENSDART00000159255 |
morn4
|
MORN repeat containing 4 |
| chr24_-_6129575 | 0.89 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr21_+_45693978 | 0.87 |
ENSDART00000160324
|
CABZ01102047.1
|
ENSDARG00000099648 |
| chr17_-_26586506 | 0.87 |
ENSDART00000087062
|
fam149b1
|
family with sequence similarity 149, member B1 |
| chr16_-_13704905 | 0.85 |
ENSDART00000060004
|
ntd5
|
ntl-dependent gene 5 |
| chr15_-_34198725 | 0.85 |
ENSDART00000170130
|
ENSDARG00000101697
|
ENSDARG00000101697 |
| chr15_-_34198695 | 0.85 |
ENSDART00000170130
|
ENSDARG00000101697
|
ENSDARG00000101697 |
| chr1_+_30974088 | 0.84 |
ENSDART00000144957
|
as3mt
|
arsenite methyltransferase |
| chr1_-_43336274 | 0.82 |
ENSDART00000160276
|
si:ch73-109d9.3
|
si:ch73-109d9.3 |
| chr7_+_15619409 | 0.82 |
ENSDART00000144765
ENSDART00000145946 |
pax6b
|
paired box 6b |
| chr10_+_39211384 | 0.82 |
|
|
|
| chr21_-_41859818 | 0.81 |
ENSDART00000139698
|
endou2
|
endonuclease, polyU-specific 2 |
| chr19_-_7069850 | 0.80 |
ENSDART00000145741
|
znf384l
|
zinc finger protein 384 like |
| chr21_+_45780315 | 0.79 |
|
|
|
| chr15_-_47260896 | 0.78 |
ENSDART00000098711
|
eif3k
|
eukaryotic translation initiation factor 3, subunit K |
| chr18_+_49230636 | 0.77 |
ENSDART00000167609
ENSDART00000135026 ENSDART00000171618 |
si:ch211-136a13.1
|
si:ch211-136a13.1 |
| chr13_-_4238224 | 0.77 |
ENSDART00000124716
|
si:dkeyp-121d4.3
|
si:dkeyp-121d4.3 |
| chr22_+_24362832 | 0.77 |
ENSDART00000157861
|
p3h2
|
prolyl 3-hydroxylase 2 |
| chr12_-_1545172 | 0.77 |
|
|
|
| chr23_+_35954132 | 0.77 |
ENSDART00000103149
|
hoxc13a
|
homeobox C13a |
| KN150135v1_+_3896 | 0.75 |
ENSDART00000174906
|
CABZ01008508.1
|
ENSDARG00000106459 |
| chr23_+_200650 | 0.74 |
|
|
|
| chr7_+_25735562 | 0.74 |
ENSDART00000112908
|
pelp1
|
proline, glutamate and leucine rich protein 1 |
| chr6_+_7395817 | 0.74 |
ENSDART00000150939
ENSDART00000151114 |
myh10
|
myosin, heavy chain 10, non-muscle |
| chr20_-_54645287 | 0.72 |
ENSDART00000153389
|
yy1b
|
YY1 transcription factor b |
| chr1_-_30772653 | 0.71 |
ENSDART00000087115
|
rims1b
|
regulating synaptic membrane exocytosis 1b |
| chr8_-_386408 | 0.70 |
|
|
|
| chr5_+_2839985 | 0.69 |
|
|
|
| chr20_+_26196386 | 0.64 |
ENSDART00000139350
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr18_-_5796337 | 0.63 |
|
|
|
| chr11_+_2786083 | 0.62 |
ENSDART00000172837
|
kif21b
|
kinesin family member 21B |
| KN150339v1_-_34304 | 0.60 |
|
|
|
| chr21_-_44707326 | 0.58 |
ENSDART00000013814
|
ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 homolog (human) |
| chr3_+_12132746 | 0.56 |
|
|
|
| chr10_-_39211281 | 0.54 |
ENSDART00000148825
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr5_-_52197945 | 0.54 |
|
|
|
| chr10_+_17490443 | 0.52 |
|
|
|
| chr7_+_10460031 | 0.52 |
ENSDART00000167323
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr10_-_37098396 | 0.50 |
ENSDART00000155277
|
BX323076.2
|
ENSDARG00000097288 |
| chr16_+_11295005 | 0.49 |
|
|
|
| chr25_+_32062428 | 0.49 |
ENSDART00000152326
|
sqrdl
|
sulfide quinone reductase-like (yeast) |
| chr5_+_35913418 | 0.47 |
ENSDART00000013590
|
wnt11
|
wingless-type MMTV integration site family, member 11 |
| chr12_-_26323696 | 0.47 |
|
|
|
| chr20_-_25727347 | 0.45 |
ENSDART00000136475
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr22_+_29694605 | 0.43 |
|
|
|
| chr7_+_20008453 | 0.42 |
ENSDART00000012450
|
dvl2
|
dishevelled segment polarity protein 2 |
| chr7_+_29680933 | 0.42 |
ENSDART00000173540
|
tpma
|
alpha-tropomyosin |
| chr20_-_54206786 | 0.40 |
ENSDART00000004756
|
hsp90aa1.1
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 1 |
| chr23_+_4287444 | 0.39 |
ENSDART00000012445
|
zgc:113278
|
zgc:113278 |
| chr7_+_29681196 | 0.39 |
ENSDART00000039657
|
tpma
|
alpha-tropomyosin |
| chr20_+_22780756 | 0.38 |
|
|
|
| chr21_-_44707514 | 0.38 |
ENSDART00000013814
|
ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 homolog (human) |
| chr2_-_59006559 | 0.38 |
|
|
|
| chr10_-_14530340 | 0.37 |
ENSDART00000101298
ENSDART00000138161 |
galt
|
galactose-1-phosphate uridylyltransferase |
| chr20_+_25727285 | 0.37 |
ENSDART00000143883
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr7_+_29681510 | 0.36 |
ENSDART00000173737
|
tpma
|
alpha-tropomyosin |
| chr8_-_16556749 | 0.35 |
ENSDART00000101655
|
calr
|
calreticulin |
| chr13_-_4238408 | 0.33 |
ENSDART00000124716
|
si:dkeyp-121d4.3
|
si:dkeyp-121d4.3 |
| chr18_-_1620953 | 0.32 |
|
|
|
| chr10_-_45535333 | 0.31 |
|
|
|
| chr18_+_18598885 | 0.31 |
|
|
|
| chr7_-_18281871 | 0.29 |
|
|
|
| chr8_+_7882144 | 0.29 |
|
|
|
| chr20_+_54645315 | 0.29 |
|
|
|
| chr21_+_17509116 | 0.27 |
|
|
|
| chr11_+_40567461 | 0.26 |
ENSDART00000160023
|
errfi1a
|
ERBB receptor feedback inhibitor 1a |
| chr9_+_24344001 | 0.26 |
|
|
|
| chr21_+_42450453 | 0.26 |
|
|
|
| chr17_-_1496133 | 0.26 |
ENSDART00000175509
|
SLC25A29
|
solute carrier family 25 member 29 |
| chr18_-_40763343 | 0.24 |
ENSDART00000130397
|
akt2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr20_-_51373292 | 0.22 |
ENSDART00000027836
ENSDART00000114407 |
rbm25b
|
RNA binding motif protein 25b |
| chr9_+_17855021 | 0.21 |
ENSDART00000166566
ENSDART00000060356 |
dgkh
|
diacylglycerol kinase, eta |
| chr5_+_35913494 | 0.19 |
ENSDART00000013590
|
wnt11
|
wingless-type MMTV integration site family, member 11 |
| chr7_+_25735735 | 0.19 |
ENSDART00000112908
|
pelp1
|
proline, glutamate and leucine rich protein 1 |
| chr5_+_35839198 | 0.18 |
ENSDART00000102973
ENSDART00000103020 |
eda
|
ectodysplasin A |
| chr7_+_10460159 | 0.16 |
ENSDART00000158162
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr21_-_37404533 | 0.15 |
ENSDART00000129439
|
BX649250.1
|
ENSDARG00000091187 |
| chr3_+_32711007 | 0.14 |
ENSDART00000122228
|
prr14
|
proline rich 14 |
| chr7_+_10459899 | 0.13 |
ENSDART00000171385
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr19_-_42668754 | 0.13 |
ENSDART00000132591
|
si:ch211-191i18.4
|
si:ch211-191i18.4 |
| chr12_-_31307581 | 0.12 |
ENSDART00000153172
|
vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chr8_-_43733891 | 0.10 |
ENSDART00000166581
|
ep400
|
E1A binding protein p400 |
| chr4_+_14659537 | 0.10 |
|
|
|
| KN149739v1_+_40107 | 0.07 |
|
|
|
| chr25_+_29631947 | 0.06 |
ENSDART00000154849
|
ENSDARG00000029431
|
ENSDARG00000029431 |
| chr18_+_58161 | 0.06 |
|
|
|
| chr14_+_28180174 | 0.05 |
ENSDART00000161852
|
stag2b
|
stromal antigen 2b |
| chr2_+_42155813 | 0.05 |
ENSDART00000143562
|
gbp2
|
guanylate binding protein 2 |
| chr20_+_14073464 | 0.04 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr23_+_18852697 | 0.03 |
ENSDART00000158267
|
emp3b
|
epithelial membrane protein 3b |
| chr11_-_21143897 | 0.02 |
ENSDART00000163008
|
RASSF5
|
Ras association domain family member 5 |
| chr24_-_39630728 | 0.01 |
ENSDART00000031486
|
lyrm1
|
LYR motif containing 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.5 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.6 | 3.7 | GO:0045428 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.6 | 4.0 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.6 | 1.7 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.5 | 2.2 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.5 | 2.2 | GO:1901492 | regulation of lymphangiogenesis(GO:1901490) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.5 | 2.8 | GO:0032099 | regulation of response to food(GO:0032095) negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.4 | 2.2 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.3 | 1.9 | GO:0050427 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.3 | 1.6 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.2 | 0.9 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.2 | 1.5 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.2 | 0.8 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.2 | 3.8 | GO:0014823 | response to activity(GO:0014823) |
| 0.2 | 0.9 | GO:1904325 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) positive regulation of calcium ion transport into cytosol(GO:0010524) positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904323) positive regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904325) |
| 0.2 | 0.8 | GO:0090104 | pancreatic epsilon cell differentiation(GO:0090104) |
| 0.2 | 1.6 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 2.3 | GO:0044259 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.1 | 1.3 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 1.5 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.8 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 2.3 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.7 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.7 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 | 1.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.3 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.8 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.4 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 1.8 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 4.9 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 1.0 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.7 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.4 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 1.0 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 2.3 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.7 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.1 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.3 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.3 | 3.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 3.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.7 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 2.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 2.3 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 1.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 2.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 2.7 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 4.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.6 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.8 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 1.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.6 | GO:0005871 | kinesin complex(GO:0005871) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.5 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.6 | 2.3 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.5 | 1.9 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.3 | 1.9 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.3 | 2.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.3 | 4.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.3 | 3.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 2.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.2 | 0.8 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.2 | 0.9 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 0.4 | GO:0004044 | amidophosphoribosyltransferase activity(GO:0004044) |
| 0.1 | 1.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.4 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.3 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 1.0 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 0.4 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 2.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 2.3 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 4.9 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 3.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.0 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.9 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 1.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.1 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 2.3 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 2.7 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.8 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.6 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 0.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.9 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 2.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.3 | 2.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 3.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.2 | 7.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 0.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |