Project
DANIO-CODE
Navigation
Downloads
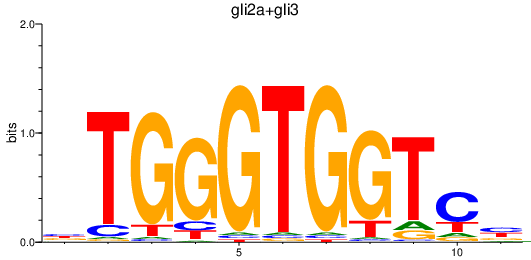
Results for gli2a+gli3
Z-value: 0.65
Transcription factors associated with gli2a+gli3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gli2a
|
ENSDARG00000025641 | GLI family zinc finger 2a |
|
gli3
|
ENSDARG00000052131 | GLI family zinc finger 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gli3 | dr10_dc_chr24_-_11681151_11681173 | 0.82 | 1.2e-04 | Click! |
| gli2a | dr10_dc_chr9_+_37331936_37332067 | 0.69 | 3.3e-03 | Click! |
Activity profile of gli2a+gli3 motif
Sorted Z-values of gli2a+gli3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of gli2a+gli3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_54876680 | 2.83 |
ENSDART00000111585
|
hbbe1.3
|
hemoglobin, beta embryonic 1.3 |
| chr23_+_20496283 | 2.45 |
ENSDART00000144047
ENSDART00000104336 |
tnnc2
|
troponin C type 2 (fast) |
| chr23_+_28656263 | 2.15 |
ENSDART00000020296
|
nadl1.2
|
neural adhesion molecule L1.2 |
| chr8_+_29953399 | 2.06 |
ENSDART00000149372
ENSDART00000007640 |
ptch1
ptch1
|
patched 1 patched 1 |
| chr1_-_19152361 | 2.00 |
ENSDART00000132958
|
grhprb
|
glyoxylate reductase/hydroxypyruvate reductase b |
| chr2_-_34010299 | 1.87 |
ENSDART00000140910
|
ptch2
|
patched 2 |
| chr21_-_23271127 | 1.69 |
ENSDART00000007806
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr7_-_8128948 | 1.62 |
ENSDART00000057101
|
aep1
|
aerolysin-like protein |
| chr8_-_47811709 | 1.60 |
ENSDART00000115050
ENSDART00000133669 |
kank3
|
KN motif and ankyrin repeat domains 3 |
| chr10_+_33227967 | 1.50 |
ENSDART00000159666
|
myl10
|
myosin, light chain 10, regulatory |
| chr9_-_8752098 | 1.46 |
|
|
|
| chr11_-_80746 | 1.44 |
|
|
|
| chr12_+_42281025 | 1.42 |
ENSDART00000167324
|
ebf3a
|
early B-cell factor 3a |
| chr24_+_18154733 | 1.38 |
ENSDART00000140994
|
tpk1
|
thiamin pyrophosphokinase 1 |
| chr16_-_38117896 | 1.37 |
ENSDART00000114266
|
plekho1b
|
pleckstrin homology domain containing, family O member 1b |
| chr22_+_38220826 | 1.33 |
ENSDART00000010537
|
cp
|
ceruloplasmin |
| chr4_+_5124186 | 1.30 |
ENSDART00000127874
|
ccnd2b
|
cyclin D2, b |
| chr20_-_39693535 | 1.29 |
ENSDART00000023531
|
hey2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr10_+_18994733 | 1.28 |
ENSDART00000146517
|
dpysl2b
|
dihydropyrimidinase-like 2b |
| chr1_+_50835265 | 1.28 |
ENSDART00000162226
|
meis1a
|
Meis homeobox 1 a |
| chr24_-_26138298 | 1.25 |
ENSDART00000128618
|
and1
|
actinodin1 |
| chr15_-_35394535 | 1.14 |
ENSDART00000144153
|
mff
|
mitochondrial fission factor |
| chr12_-_17357133 | 1.09 |
ENSDART00000079115
|
papss2b
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2b |
| chr12_+_27026112 | 1.09 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr11_+_36983626 | 1.08 |
ENSDART00000170209
|
il17rc
|
interleukin 17 receptor C |
| chr9_-_46272322 | 1.04 |
ENSDART00000169682
|
hdac4
|
histone deacetylase 4 |
| chr13_+_30673907 | 1.03 |
ENSDART00000053946
|
cxcl12a
|
chemokine (C-X-C motif) ligand 12a (stromal cell-derived factor 1) |
| chr5_+_32732535 | 1.02 |
ENSDART00000156913
|
BX927184.1
|
ENSDARG00000097904 |
| chr20_+_15653121 | 0.98 |
ENSDART00000152702
|
jun
|
jun proto-oncogene |
| chr6_+_6967174 | 0.97 |
ENSDART00000151311
|
ENSDARG00000058903
|
ENSDARG00000058903 |
| chr7_-_38288929 | 0.95 |
|
|
|
| chr23_+_4475256 | 0.92 |
ENSDART00000081821
|
wnt7ab
|
wingless-type MMTV integration site family, member 7Ab |
| chr16_-_5006929 | 0.92 |
ENSDART00000054077
|
tpbgb
|
trophoblast glycoprotein b |
| chr22_+_1568413 | 0.91 |
ENSDART00000175704
|
si:ch211-255f4.11
|
si:ch211-255f4.11 |
| chr19_-_19282688 | 0.88 |
ENSDART00000162708
|
ppt2
|
palmitoyl-protein thioesterase 2 |
| chr9_+_8752130 | 0.88 |
ENSDART00000137813
|
FP103004.1
|
ENSDARG00000092413 |
| chr16_-_7963894 | 0.87 |
ENSDART00000172836
|
CABZ01061349.1
|
ENSDARG00000105487 |
| chr1_+_3674819 | 0.78 |
|
|
|
| chr12_-_15524097 | 0.76 |
|
|
|
| chr13_+_41998500 | 0.73 |
ENSDART00000074707
|
cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr16_+_5466342 | 0.73 |
ENSDART00000160008
|
plecb
|
plectin b |
| chr12_-_1545172 | 0.73 |
|
|
|
| chr20_-_28739099 | 0.71 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr19_-_10941519 | 0.71 |
ENSDART00000169118
ENSDART00000160599 |
CU855694.1
|
ENSDARG00000098513 |
| chr2_-_59006559 | 0.69 |
|
|
|
| chr10_-_1799784 | 0.68 |
ENSDART00000058627
|
epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr8_+_47352102 | 0.66 |
|
|
|
| chr14_-_7772 | 0.63 |
ENSDART00000054822
|
nkx3.2
|
NK3 homeobox 2 |
| chr16_-_43090120 | 0.63 |
ENSDART00000155637
|
BX005234.1
|
ENSDARG00000097761 |
| chr19_-_10324632 | 0.62 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr17_+_45931793 | 0.61 |
ENSDART00000155372
|
kif26ab
|
kinesin family member 26Ab |
| chr9_-_10561062 | 0.60 |
ENSDART00000175269
|
thsd7ba
|
thrombospondin, type I, domain containing 7Ba |
| chr10_+_26786051 | 0.60 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr24_-_35486496 | 0.59 |
|
|
|
| chr2_+_47864913 | 0.59 |
|
|
|
| chr25_+_1773157 | 0.58 |
|
|
|
| chr10_-_15090647 | 0.58 |
ENSDART00000038401
ENSDART00000155674 |
si:ch211-95j8.2
|
si:ch211-95j8.2 |
| chr9_-_1983772 | 0.57 |
ENSDART00000082339
|
hoxd12a
|
homeobox D12a |
| chr14_-_16171165 | 0.57 |
ENSDART00000089021
|
canx
|
calnexin |
| chr8_-_14446883 | 0.55 |
ENSDART00000057644
|
lhx4
|
LIM homeobox 4 |
| chr12_-_1545141 | 0.52 |
|
|
|
| chr1_+_51712192 | 0.52 |
|
|
|
| chr6_+_7395817 | 0.52 |
ENSDART00000150939
ENSDART00000151114 |
myh10
|
myosin, heavy chain 10, non-muscle |
| chr17_-_35004687 | 0.49 |
|
|
|
| chr6_+_9192009 | 0.49 |
ENSDART00000022620
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr8_+_31110356 | 0.48 |
ENSDART00000136578
|
syn1
|
synapsin I |
| chr21_-_38571536 | 0.47 |
ENSDART00000139178
ENSDART00000036600 |
slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr21_-_30378764 | 0.47 |
ENSDART00000136247
|
grpel2
|
GrpE-like 2, mitochondrial |
| chr25_+_32062428 | 0.46 |
ENSDART00000152326
|
sqrdl
|
sulfide quinone reductase-like (yeast) |
| chr5_-_71505131 | 0.46 |
ENSDART00000037691
|
tbx5a
|
T-box 5a |
| chr18_-_1577633 | 0.45 |
|
|
|
| chr7_-_15164722 | 0.45 |
ENSDART00000172147
|
ENSDARG00000036809
|
ENSDARG00000036809 |
| chr2_-_54028761 | 0.45 |
ENSDART00000178289
|
CABZ01050256.1
|
ENSDARG00000109075 |
| chr20_+_13507202 | 0.44 |
ENSDART00000127350
|
enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr3_+_56507117 | 0.42 |
ENSDART00000154405
|
rac1b
|
ras-related C3 botulinum toxin substrate 1b (rho family, small GTP binding protein Rac1) |
| chr23_-_41929027 | 0.41 |
ENSDART00000131235
|
GMEB2
|
glucocorticoid modulatory element binding protein 2 |
| chr2_-_24947660 | 0.41 |
ENSDART00000113356
ENSDART00000163038 |
crtc1a
|
CREB regulated transcription coactivator 1a |
| chr13_-_40600924 | 0.41 |
ENSDART00000099847
ENSDART00000057046 |
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr14_+_50164568 | 0.40 |
ENSDART00000084114
|
nsd1a
|
nuclear receptor binding SET domain protein 1a |
| KN150339v1_-_34304 | 0.40 |
|
|
|
| chr9_+_6609374 | 0.39 |
ENSDART00000122279
|
fhl2a
|
four and a half LIM domains 2a |
| chr9_-_8752195 | 0.38 |
|
|
|
| chr2_+_27000558 | 0.38 |
ENSDART00000099208
|
asph
|
aspartate beta-hydroxylase |
| chr15_+_2590598 | 0.38 |
ENSDART00000178505
|
SH2B2
|
SH2B adaptor protein 2 |
| chr2_-_6139895 | 0.37 |
ENSDART00000147509
|
si:ch211-284b7.3
|
si:ch211-284b7.3 |
| chr20_+_22780756 | 0.34 |
|
|
|
| chr18_+_20504641 | 0.33 |
|
|
|
| chr20_+_26196386 | 0.33 |
ENSDART00000139350
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr22_-_11024472 | 0.32 |
ENSDART00000105823
ENSDART00000159995 |
insrb
|
insulin receptor b |
| chr7_+_24983985 | 0.31 |
|
|
|
| chr24_-_38758093 | 0.31 |
|
|
|
| chr18_+_54352 | 0.31 |
ENSDART00000097163
|
SLC27A2 (1 of many)
|
solute carrier family 27 member 2 |
| chr13_+_41998172 | 0.29 |
ENSDART00000074707
|
cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr7_-_18281871 | 0.28 |
|
|
|
| chr13_+_41998364 | 0.28 |
ENSDART00000074707
|
cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr8_+_28433869 | 0.27 |
ENSDART00000062716
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr14_+_43599635 | 0.27 |
ENSDART00000155539
|
CR786577.1
|
ENSDARG00000097875 |
| chr9_-_11683760 | 0.26 |
ENSDART00000044314
ENSDART00000124129 |
itgav
|
integrin, alpha V |
| chr5_+_32732654 | 0.26 |
ENSDART00000156913
|
BX927184.1
|
ENSDARG00000097904 |
| chr7_+_25735562 | 0.25 |
ENSDART00000112908
|
pelp1
|
proline, glutamate and leucine rich protein 1 |
| chr16_+_37519702 | 0.25 |
|
|
|
| chr14_-_32542387 | 0.22 |
|
|
|
| chr24_-_25528893 | 0.22 |
ENSDART00000142351
|
cnksr2a
|
connector enhancer of kinase suppressor of Ras 2a |
| chr16_+_37519812 | 0.21 |
|
|
|
| chr1_-_49144795 | 0.20 |
|
|
|
| chr25_+_7287952 | 0.19 |
ENSDART00000170569
|
syt12
|
synaptotagmin XII |
| chr6_-_47928632 | 0.18 |
|
|
|
| chr7_+_20008453 | 0.18 |
ENSDART00000012450
|
dvl2
|
dishevelled segment polarity protein 2 |
| chr20_+_25727285 | 0.16 |
ENSDART00000143883
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr24_+_17190237 | 0.15 |
ENSDART00000126435
|
pdia4
|
protein disulfide isomerase family A, member 4 |
| chr20_-_6087735 | 0.15 |
ENSDART00000145964
|
pum2
|
pumilio RNA-binding family member 2 |
| chr17_+_45931756 | 0.15 |
ENSDART00000155372
|
kif26ab
|
kinesin family member 26Ab |
| chr21_+_11686197 | 0.15 |
ENSDART00000031786
|
glrx
|
glutaredoxin (thioltransferase) |
| chr7_+_51541162 | 0.14 |
ENSDART00000174120
|
si:ch211-122f10.4
|
si:ch211-122f10.4 |
| chr20_-_25727347 | 0.12 |
ENSDART00000136475
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr19_-_11480368 | 0.12 |
ENSDART00000080762
|
sept7a
|
septin 7a |
| chr17_-_30958965 | 0.11 |
ENSDART00000051697
|
evla
|
Enah/Vasp-like a |
| chr20_+_4530154 | 0.10 |
ENSDART00000168322
|
tiam2b
|
T-cell lymphoma invasion and metastasis 2b |
| chr17_+_37354614 | 0.10 |
|
|
|
| chr19_+_27094665 | 0.08 |
ENSDART00000079410
|
si:dkey-27c15.3
|
si:dkey-27c15.3 |
| chr24_+_7799283 | 0.05 |
ENSDART00000135161
|
zgc:101569
|
zgc:101569 |
| chr17_+_37354528 | 0.04 |
|
|
|
| chr1_+_44971899 | 0.03 |
ENSDART00000149565
|
trappc5
|
trafficking protein particle complex 5 |
| chr16_+_42763696 | 0.01 |
ENSDART00000023452
|
dpy19l1l
|
dpy-19-like 1, like (H. sapiens) |
| chr5_-_52357382 | 0.00 |
ENSDART00000157978
|
mrps30
|
mitochondrial ribosomal protein S30 |
| chr16_+_41923717 | 0.00 |
ENSDART00000084631
|
scn1ba
|
sodium channel, voltage-gated, type I, beta a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.9 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.5 | 1.4 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine diphosphate metabolic process(GO:0042357) thiamine-containing compound metabolic process(GO:0042723) |
| 0.4 | 1.3 | GO:0060853 | Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) |
| 0.4 | 1.7 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.3 | 1.0 | GO:0036073 | perichondral ossification(GO:0036073) |
| 0.3 | 1.0 | GO:2000471 | negative regulation of leukocyte migration(GO:0002686) regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.3 | 1.4 | GO:1901739 | regulation of myoblast fusion(GO:1901739) |
| 0.3 | 2.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 1.1 | GO:0050427 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 | 2.4 | GO:0014823 | response to activity(GO:0014823) |
| 0.2 | 0.5 | GO:0003294 | atrial ventricular junction remodeling(GO:0003294) |
| 0.1 | 0.4 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 1.3 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 0.4 | GO:0055062 | regulation of fat cell differentiation(GO:0045598) phosphate ion homeostasis(GO:0055062) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.1 | 1.2 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.6 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 1.3 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.5 | GO:0015743 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.1 | 0.6 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 1.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.7 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 0.5 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 1.6 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.2 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 1.6 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.2 | GO:0090177 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.3 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.6 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.7 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.3 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.6 | GO:0007596 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.0 | 1.3 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 1.2 | GO:0033333 | fin development(GO:0033333) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.8 | GO:0005833 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 0.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 1.6 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 0.5 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 2.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.5 | GO:0016459 | myosin complex(GO:0016459) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | GO:0008158 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.7 | 2.0 | GO:0008465 | glycerate dehydrogenase activity(GO:0008465) |
| 0.4 | 2.8 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.3 | 1.6 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.3 | 1.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 1.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 1.1 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.2 | 1.4 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.2 | 1.3 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 2.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.4 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.1 | 1.0 | GO:0042379 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.1 | 0.5 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) succinate transmembrane transporter activity(GO:0015141) |
| 0.1 | 1.0 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.4 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.1 | 0.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.2 | GO:0004044 | amidophosphoribosyltransferase activity(GO:0004044) |
| 0.0 | 0.3 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.7 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.0 | 0.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.3 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.5 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.1 | 3.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 0.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.3 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 2.3 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |