Project
DANIO-CODE
Navigation
Downloads
Results for glis2a+glis2b
Z-value: 0.91
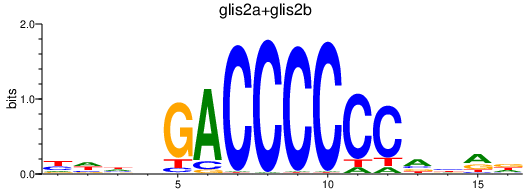
Transcription factors associated with glis2a+glis2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
glis2a
|
ENSDARG00000078388 | GLIS family zinc finger 2a |
|
glis2b
|
ENSDARG00000100232 | GLIS family zinc finger 2b |
Activity profile of glis2a+glis2b motif
Sorted Z-values of glis2a+glis2b motif
Network of associatons between targets according to the STRING database.
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_31672763 | 3.49 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr9_-_23081250 | 3.24 |
|
|
|
| chr7_+_57423271 | 3.05 |
ENSDART00000056466
|
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr16_+_4825019 | 3.00 |
ENSDART00000167665
|
LIN28A
|
lin-28 homolog A |
| chr7_+_18111890 | 2.87 |
ENSDART00000171606
|
cd248a
|
CD248 molecule, endosialin a |
| chr4_-_24310846 | 2.57 |
ENSDART00000017443
|
celf2
|
cugbp, Elav-like family member 2 |
| chr4_-_17735989 | 2.54 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr10_+_44364117 | 2.20 |
|
|
|
| chr7_+_11148688 | 2.15 |
ENSDART00000114383
|
mesdc1
|
mesoderm development candidate 1 |
| chr15_-_22881 | 2.10 |
ENSDART00000158734
|
BACE1
|
beta-secretase 1 |
| chr12_+_9842842 | 2.03 |
ENSDART00000055019
|
ndufa4
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4 |
| chr15_-_22978 | 1.93 |
ENSDART00000158734
|
BACE1
|
beta-secretase 1 |
| chr20_+_25441689 | 1.89 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr9_-_6949104 | 1.88 |
ENSDART00000059092
|
tmem182a
|
transmembrane protein 182a |
| chr16_-_16682692 | 1.69 |
ENSDART00000059841
|
si:ch211-257p13.3
|
si:ch211-257p13.3 |
| chr12_-_41124276 | 1.68 |
ENSDART00000172022
ENSDART00000158605 ENSDART00000162967 |
dpysl4
|
dihydropyrimidinase-like 4 |
| chr13_-_508050 | 1.54 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr13_-_508202 | 1.50 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr20_-_1218693 | 1.47 |
ENSDART00000140650
ENSDART00000133216 |
ankrd6b
|
ankyrin repeat domain 6b |
| chr5_-_41672394 | 1.40 |
ENSDART00000164363
|
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr23_-_17543731 | 1.31 |
ENSDART00000002398
|
trim101
|
tripartite motif containing 101 |
| chr2_+_46428154 | 1.29 |
ENSDART00000128457
|
ephb1
|
EPH receptor B1 |
| chr3_+_21059221 | 1.21 |
ENSDART00000078807
|
zgc:123295
|
zgc:123295 |
| chr24_+_25326286 | 1.17 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr5_-_21497169 | 1.10 |
ENSDART00000143878
|
arhgef9a
|
Cdc42 guanine nucleotide exchange factor (GEF) 9a |
| chr8_+_6989445 | 1.09 |
ENSDART00000134440
|
gpd1a
|
glycerol-3-phosphate dehydrogenase 1a |
| chr24_+_25326233 | 0.99 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr11_+_27721622 | 0.94 |
ENSDART00000147984
|
alpl
|
alkaline phosphatase, liver/bone/kidney |
| KN150222v1_+_74557 | 0.92 |
|
|
|
| chr24_-_9819862 | 0.91 |
ENSDART00000092975
|
vps41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr23_+_24778062 | 0.86 |
|
|
|
| chr5_+_50432868 | 0.85 |
ENSDART00000097466
|
fam169aa
|
family with sequence similarity 169, member Aa |
| chr8_-_13182911 | 0.84 |
ENSDART00000090541
|
si:ch73-61d6.3
|
si:ch73-61d6.3 |
| chr24_-_25545773 | 0.68 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr9_+_8131114 | 0.68 |
|
|
|
| chr2_+_14039168 | 0.63 |
ENSDART00000155015
|
zbtb41
|
zinc finger and BTB domain containing 41 |
| chr8_-_13183034 | 0.62 |
ENSDART00000090541
|
si:ch73-61d6.3
|
si:ch73-61d6.3 |
| chr16_+_5339396 | 0.60 |
|
|
|
| chr16_+_4825665 | 0.59 |
|
|
|
| chr7_-_19390325 | 0.54 |
ENSDART00000163686
|
si:ch211-212k18.4
|
si:ch211-212k18.4 |
| chr19_-_43358940 | 0.53 |
|
|
|
| chr12_-_44825493 | 0.51 |
ENSDART00000016635
|
bccip
|
BRCA2 and CDKN1A interacting protein |
| chr9_-_2416300 | 0.51 |
|
|
|
| chr19_-_43359483 | 0.48 |
ENSDART00000086659
|
cyhr1
|
cysteine/histidine-rich 1 |
| chr7_+_48431214 | 0.48 |
|
|
|
| chr23_+_24778090 | 0.48 |
|
|
|
| chr16_-_45888127 | 0.40 |
ENSDART00000027013
ENSDART00000128068 |
ntrk1
|
neurotrophic tyrosine kinase, receptor, type 1 |
| chr12_-_196397 | 0.37 |
ENSDART00000161178
ENSDART00000160926 |
dnah9
|
dynein, axonemal, heavy chain 9 |
| chr17_+_23558463 | 0.35 |
ENSDART00000149281
|
slc16a12a
|
solute carrier family 16, member 12a |
| chr11_+_27721554 | 0.29 |
ENSDART00000131101
|
alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr17_-_23654438 | 0.28 |
ENSDART00000122209
|
ptena
|
phosphatase and tensin homolog A |
| chr10_-_13158018 | 0.26 |
ENSDART00000164568
|
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr12_-_1565781 | 0.25 |
ENSDART00000170055
|
CABZ01073190.1
|
ENSDARG00000100999 |
| chr10_+_44364064 | 0.24 |
|
|
|
| chr12_+_31388957 | 0.23 |
|
|
|
| chr13_-_508110 | 0.22 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr1_-_53465275 | 0.14 |
|
|
|
| chr24_-_35819359 | 0.12 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr3_-_61087420 | 0.11 |
|
|
|
| chr16_+_25891403 | 0.08 |
ENSDART00000154652
|
irgq1
|
immunity-related GTPase family, q1 |
| chr15_+_557244 | 0.06 |
ENSDART00000154594
ENSDART00000102268 |
pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.2 | 1.1 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.2 | 0.9 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.2 | 1.7 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 0.4 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.1 | 2.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.3 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.1 | 1.5 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.3 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 1.5 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 1.2 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.0 | 0.5 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.4 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 2.5 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.1 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 1.7 | GO:0007018 | microtubule-based movement(GO:0007018) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 0.9 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.2 | 1.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 3.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.5 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.1 | GO:0014069 | postsynaptic density(GO:0014069) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.8 | 2.5 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.4 | 3.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 1.7 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 1.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 3.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.4 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 1.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.3 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.1 | 0.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.3 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 1.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 2.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 1.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.4 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 2.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 3.5 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 0.7 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.1 | 0.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 2.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |