Project
DANIO-CODE
Navigation
Downloads
Results for glis3
Z-value: 0.88
Transcription factors associated with glis3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
glis3
|
ENSDARG00000069726 | GLIS family zinc finger 3 |
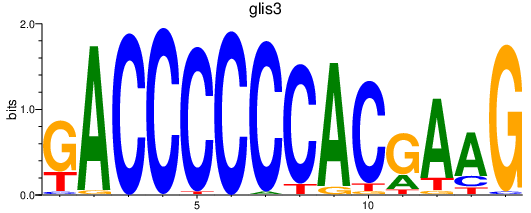
Activity profile of glis3 motif
Sorted Z-values of glis3 motif
Network of associatons between targets according to the STRING database.
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_30363703 | 3.73 |
ENSDART00000134098
|
efemp2a
|
EGF containing fibulin-like extracellular matrix protein 2a |
| chr6_-_22504772 | 3.58 |
ENSDART00000170039
|
sept9b
|
septin 9b |
| chr15_-_41288480 | 2.51 |
ENSDART00000155359
|
smco4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr8_-_15144533 | 2.34 |
ENSDART00000138855
|
bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr8_+_13064944 | 2.11 |
|
|
|
| chr13_-_508202 | 2.00 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr8_+_46651079 | 1.87 |
ENSDART00000113803
|
her3
|
hairy-related 3 |
| chr13_-_508050 | 1.75 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr18_+_7350133 | 1.52 |
ENSDART00000172404
|
cers3b
|
ceramide synthase 3b |
| chr18_+_9228336 | 1.13 |
|
|
|
| chr18_-_41384938 | 0.92 |
ENSDART00000098673
|
ptx3a
|
pentraxin 3, long a |
| chr14_-_33704021 | 0.70 |
ENSDART00000149396
ENSDART00000123607 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr5_+_66984082 | 0.65 |
ENSDART00000137700
|
si:dkey-70b23.2
|
si:dkey-70b23.2 |
| chr13_+_28723589 | 0.50 |
|
|
|
| chr14_-_28228583 | 0.46 |
ENSDART00000054088
|
zgc:113364
|
zgc:113364 |
| chr3_+_30059565 | 0.42 |
ENSDART00000157320
|
akt1s1
|
AKT1 substrate 1 (proline-rich) |
| chr17_+_23558463 | 0.38 |
ENSDART00000149281
|
slc16a12a
|
solute carrier family 16, member 12a |
| chr25_+_7287952 | 0.26 |
ENSDART00000170569
|
syt12
|
synaptotagmin XII |
| chr13_-_508110 | 0.23 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.3 | 4.0 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.2 | 2.3 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 0.4 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 0.7 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 3.6 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.3 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 1.5 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 1.9 | GO:0045665 | negative regulation of neuron differentiation(GO:0045665) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.6 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 8.6 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 1.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 3.6 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.4 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 2.3 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.4 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |