Project
DANIO-CODE
Navigation
Downloads
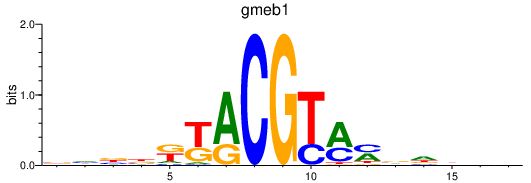
Results for gmeb1
Z-value: 0.40
Transcription factors associated with gmeb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
gmeb1
|
ENSDARG00000062060 | glucocorticoid modulatory element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| gmeb1 | dr10_dc_chr19_+_16159980_16160000 | 0.87 | 1.1e-05 | Click! |
Activity profile of gmeb1 motif
Sorted Z-values of gmeb1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of gmeb1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_2296253 | 1.49 |
ENSDART00000162867
|
zgc:66483
|
zgc:66483 |
| chr14_-_31149030 | 0.99 |
ENSDART00000018347
|
cab39l1
|
calcium binding protein 39, like 1 |
| chr22_+_24596299 | 0.95 |
ENSDART00000158303
ENSDART00000160924 |
mcoln2
|
mucolipin 2 |
| chr16_-_41585481 | 0.93 |
ENSDART00000102662
|
rpp25l
|
ribonuclease P/MRP 25 subunit-like |
| chr12_-_2834771 | 0.73 |
ENSDART00000114854
ENSDART00000163759 |
zdhhc16b
|
zinc finger, DHHC-type containing 16b |
| chr14_+_31149207 | 0.60 |
|
|
|
| chr13_+_29904913 | 0.54 |
ENSDART00000057525
|
dnajb12a
|
DnaJ (Hsp40) homolog, subfamily B, member 12a |
| chr4_+_17653548 | 0.50 |
ENSDART00000026509
|
cwf19l1
|
CWF19-like 1, cell cycle control |
| chr25_-_34340016 | 0.50 |
ENSDART00000039485
|
gabarapl2
|
GABA(A) receptor-associated protein like 2 |
| chr23_-_9872524 | 0.49 |
ENSDART00000133602
|
mapre1b
|
microtubule-associated protein, RP/EB family, member 1b |
| chr16_-_41585555 | 0.41 |
ENSDART00000131706
|
rpp25l
|
ribonuclease P/MRP 25 subunit-like |
| chr25_+_22183891 | 0.40 |
ENSDART00000089516
|
stoml1
|
stomatin (EPB72)-like 1 |
| chr10_+_29251234 | 0.40 |
ENSDART00000123033
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr12_-_2834814 | 0.39 |
ENSDART00000114854
ENSDART00000163759 |
zdhhc16b
|
zinc finger, DHHC-type containing 16b |
| chr14_-_44794907 | 0.37 |
ENSDART00000172805
|
CABZ01074913.1
|
ENSDARG00000105401 |
| chr3_+_32802124 | 0.36 |
ENSDART00000112742
|
nbr1b
|
neighbor of brca1 gene 1b |
| chr17_-_25813136 | 0.35 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr3_+_48811909 | 0.34 |
ENSDART00000136051
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr12_-_48391342 | 0.30 |
ENSDART00000153209
|
dnajb12b
|
DnaJ (Hsp40) homolog, subfamily B, member 12b |
| chr3_+_32801936 | 0.25 |
ENSDART00000112742
|
nbr1b
|
neighbor of brca1 gene 1b |
| chr14_-_44794961 | 0.24 |
ENSDART00000172805
|
CABZ01074913.1
|
ENSDARG00000105401 |
| chr3_+_48811662 | 0.24 |
ENSDART00000023814
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr10_+_29251158 | 0.23 |
ENSDART00000123033
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr22_+_22412894 | 0.22 |
ENSDART00000089622
|
kif14
|
kinesin family member 14 |
| chr4_+_17653739 | 0.21 |
ENSDART00000026509
|
cwf19l1
|
CWF19-like 1, cell cycle control |
| chr5_-_67315284 | 0.20 |
ENSDART00000131665
|
gtf3aa
|
general transcription factor IIIAa |
| chr5_+_29988157 | 0.15 |
ENSDART00000086661
|
dpagt1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr17_+_43478181 | 0.15 |
ENSDART00000055487
|
chmp3
|
charged multivesicular body protein 3 |
| chr9_+_16304358 | 0.13 |
ENSDART00000154326
|
si:ch211-261p9.4
|
si:ch211-261p9.4 |
| chr10_+_25234636 | 0.13 |
ENSDART00000024514
|
chordc1b
|
cysteine and histidine-rich domain (CHORD) containing 1b |
| chr23_-_9872554 | 0.11 |
ENSDART00000133602
|
mapre1b
|
microtubule-associated protein, RP/EB family, member 1b |
| chr25_+_35540123 | 0.10 |
ENSDART00000126326
|
rpgrip1l
|
RPGRIP1-like |
| chr25_-_28225408 | 0.09 |
ENSDART00000126490
|
fnbp4
|
formin binding protein 4 |
| chr7_-_18916067 | 0.08 |
ENSDART00000112447
|
il13ra1
|
interleukin 13 receptor, alpha 1 |
| chr4_-_71875028 | 0.08 |
ENSDART00000133274
|
si:ch73-266f23.1
|
si:ch73-266f23.1 |
| chr25_+_18613868 | 0.07 |
ENSDART00000011149
|
fam185a
|
family with sequence similarity 185, member A |
| chr13_-_12353513 | 0.07 |
ENSDART00000134132
|
lamtor3
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
| chr12_+_11611780 | 0.06 |
ENSDART00000150191
|
waplb
|
WAPL cohesin release factor b |
| chr4_+_23404697 | 0.05 |
ENSDART00000077854
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr21_+_26696570 | 0.03 |
ENSDART00000171641
|
pcxa
|
pyruvate carboxylase a |
| chr12_-_3630622 | 0.03 |
ENSDART00000164707
|
coa3a
|
cytochrome C oxidase assembly factor 3a |
| chr25_-_28225449 | 0.03 |
ENSDART00000126490
|
fnbp4
|
formin binding protein 4 |
| chr14_-_21635094 | 0.02 |
ENSDART00000054420
|
rad9a
|
RAD9 checkpoint clamp component A |
| chr10_-_27028132 | 0.02 |
ENSDART00000113353
|
ENSDARG00000075422
|
ENSDARG00000075422 |
| chr18_+_41244272 | 0.02 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.6 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.6 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.8 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 1.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:1904019 | epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) |
| 0.0 | 0.1 | GO:0007063 | regulation of sister chromatid cohesion(GO:0007063) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.6 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 0.6 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.7 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.6 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.1 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |