Project
DANIO-CODE
Navigation
Downloads
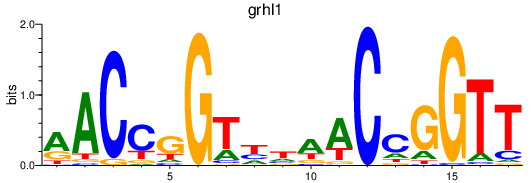
Results for grhl1
Z-value: 2.01
Transcription factors associated with grhl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
grhl1
|
ENSDARG00000061391 | grainyhead-like transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| grhl1 | dr10_dc_chr17_-_32473712_32473913 | 0.86 | 1.8e-05 | Click! |
Activity profile of grhl1 motif
Sorted Z-values of grhl1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of grhl1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_22386456 | 7.37 |
ENSDART00000146081
|
ponzr5
|
plac8 onzin related protein 5 |
| chr6_+_17959219 | 6.62 |
ENSDART00000026448
|
evpla
|
envoplakin a |
| chr3_-_29846530 | 6.43 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr24_+_38783264 | 5.62 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr9_-_30453625 | 5.06 |
ENSDART00000060150
|
mid1ip1a
|
MID1 interacting protein 1a |
| chr16_-_9785057 | 4.65 |
ENSDART00000113724
|
mal2
|
mal, T-cell differentiation protein 2 (gene/pseudogene) |
| chr6_-_43273456 | 4.63 |
|
|
|
| chr16_+_13534390 | 4.28 |
ENSDART00000157396
|
ENSDARG00000022807
|
ENSDARG00000022807 |
| KN149859v1_+_16751 | 4.18 |
ENSDART00000167610
|
ENSDARG00000103399
|
ENSDARG00000103399 |
| chr22_+_21996941 | 4.05 |
ENSDART00000149586
|
gna15.1
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 1 |
| chr21_-_22510751 | 3.89 |
ENSDART00000169870
|
myo5b
|
myosin VB |
| chr9_-_40212864 | 3.88 |
ENSDART00000166918
|
IKZF2
|
IKAROS family zinc finger 2 |
| chr16_+_7213011 | 3.71 |
ENSDART00000168830
ENSDART00000168274 ENSDART00000160383 ENSDART00000163281 |
bmper
|
BMP binding endothelial regulator |
| chr4_-_76620869 | 3.64 |
|
|
|
| chr25_+_24520476 | 3.62 |
|
|
|
| chr1_-_51862897 | 3.50 |
ENSDART00000136469
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr14_+_37206949 | 3.49 |
|
|
|
| chr4_-_876235 | 3.25 |
|
|
|
| chr1_-_23765358 | 3.24 |
|
|
|
| chr21_-_13571753 | 3.19 |
ENSDART00000065819
|
clic3
|
chloride intracellular channel 3 |
| chr3_-_18605846 | 3.09 |
ENSDART00000145277
|
zgc:113333
|
zgc:113333 |
| chr24_-_38782898 | 3.03 |
ENSDART00000155542
|
FP102170.1
|
ENSDARG00000097123 |
| chr16_+_7213161 | 3.03 |
ENSDART00000168830
ENSDART00000168274 ENSDART00000160383 ENSDART00000163281 |
bmper
|
BMP binding endothelial regulator |
| chr10_+_4924065 | 2.59 |
ENSDART00000108595
|
slc46a2
|
solute carrier family 46 member 2 |
| chr21_-_3548719 | 2.55 |
ENSDART00000137844
|
atp8b1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr4_-_9779763 | 2.55 |
ENSDART00000134280
|
svopl
|
SVOP-like |
| chr21_+_25588388 | 2.41 |
ENSDART00000134678
|
ENSDARG00000078256
|
ENSDARG00000078256 |
| chr7_+_22386328 | 2.31 |
ENSDART00000141048
|
ponzr5
|
plac8 onzin related protein 5 |
| chr13_+_18401965 | 2.22 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr4_+_4223606 | 2.05 |
|
|
|
| chr2_+_35871754 | 2.02 |
ENSDART00000134918
|
dhx9
|
DEAH (Asp-Glu-Ala-His) box helicase 9 |
| chr23_+_17428740 | 1.84 |
|
|
|
| chr1_-_51863187 | 1.81 |
ENSDART00000004233
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr5_-_55745399 | 1.71 |
|
|
|
| chr14_-_33604914 | 1.68 |
ENSDART00000168546
|
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr5_-_6004819 | 1.54 |
ENSDART00000099570
|
tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr9_-_40212643 | 1.53 |
ENSDART00000166918
|
IKZF2
|
IKAROS family zinc finger 2 |
| chr6_-_43273663 | 1.51 |
|
|
|
| chr7_+_19583578 | 1.31 |
ENSDART00000149812
|
ovol1
|
ovo-like zinc finger 1 |
| chr10_+_4924008 | 1.31 |
ENSDART00000108595
|
slc46a2
|
solute carrier family 46 member 2 |
| chr23_+_17428538 | 1.13 |
|
|
|
| chr2_-_57502941 | 0.97 |
|
|
|
| chr2_-_58272475 | 0.86 |
ENSDART00000166282
ENSDART00000159040 |
pnp5b
|
purine nucleoside phosphorylase 5b |
| chr14_+_4169846 | 0.85 |
ENSDART00000038301
|
gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr5_-_57016269 | 0.83 |
ENSDART00000074264
|
crfb12
|
cytokine receptor family member B12 |
| chr15_-_5913508 | 0.79 |
ENSDART00000155252
|
si:ch73-281n10.2
|
si:ch73-281n10.2 |
| chr5_-_39440105 | 0.77 |
ENSDART00000157755
|
cds1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr9_-_40213104 | 0.72 |
ENSDART00000166918
|
IKZF2
|
IKAROS family zinc finger 2 |
| chr16_+_7213299 | 0.71 |
ENSDART00000162610
|
bmper
|
BMP binding endothelial regulator |
| chr15_+_29343644 | 0.69 |
ENSDART00000170537
ENSDART00000128973 |
rap1gap2a
|
RAP1 GTPase activating protein 2a |
| chr8_-_49227143 | 0.67 |
|
|
|
| chr15_+_5914095 | 0.60 |
ENSDART00000114134
|
wrb
|
tryptophan rich basic protein |
| chr5_-_28970034 | 0.52 |
ENSDART00000043259
|
entpd2a.2
|
ectonucleoside triphosphate diphosphohydrolase 2a, tandem duplicate 2 |
| chr16_+_35708856 | 0.47 |
ENSDART00000161393
|
map7d1a
|
MAP7 domain containing 1a |
| chr12_-_17013870 | 0.42 |
|
|
|
| chr21_-_42813859 | 0.39 |
|
|
|
| chr20_-_43448899 | 0.39 |
ENSDART00000152888
|
mllt4a
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 4a |
| chr3_-_29846789 | 0.38 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr7_+_22421454 | 0.33 |
|
|
|
| chr9_-_40213039 | 0.31 |
ENSDART00000166918
|
IKZF2
|
IKAROS family zinc finger 2 |
| chr19_-_41887040 | 0.28 |
ENSDART00000062026
|
dlx5a
|
distal-less homeobox 5a |
| chr10_+_19568414 | 0.26 |
ENSDART00000162912
|
vsig8a
|
V-set and immunoglobulin domain containing 8a |
| chr23_-_22723966 | 0.19 |
ENSDART00000141127
|
ca6
|
carbonic anhydrase VI |
| chr14_+_4169371 | 0.19 |
ENSDART00000136665
|
gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr9_-_30453581 | 0.13 |
ENSDART00000060150
|
mid1ip1a
|
MID1 interacting protein 1a |
| chr23_-_22724076 | 0.10 |
ENSDART00000132733
|
ca6
|
carbonic anhydrase VI |
| chr19_-_20203902 | 0.08 |
ENSDART00000172668
|
CR382300.3
|
ENSDARG00000101330 |
| chr1_-_51863300 | 0.06 |
ENSDART00000004233
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr4_-_9779987 | 0.05 |
ENSDART00000134280
|
svopl
|
SVOP-like |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.8 | 4.6 | GO:0045056 | transcytosis(GO:0045056) |
| 0.4 | 4.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 | 7.5 | GO:0043049 | neural crest formation(GO:0014029) otic placode formation(GO:0043049) |
| 0.2 | 6.6 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.2 | 1.0 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.1 | 4.3 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.1 | 0.5 | GO:0071405 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.1 | 0.7 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 2.0 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.3 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 2.1 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.8 | GO:0007599 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.0 | 0.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.3 | GO:0036269 | swimming behavior(GO:0036269) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 4.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 6.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 5.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 3.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 4.6 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 7.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 4.3 | GO:0005874 | microtubule(GO:0005874) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 1.3 | 6.6 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.5 | 2.6 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.5 | 4.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.5 | 4.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 7.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 0.9 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.2 | 3.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 1.0 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 0.7 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 2.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 3.2 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 0.5 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.9 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.1 | 1.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 2.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |